MacroAPE 1083:SP4 f1
From FANTOM5_SSTAR
Full Name: SP4_f1
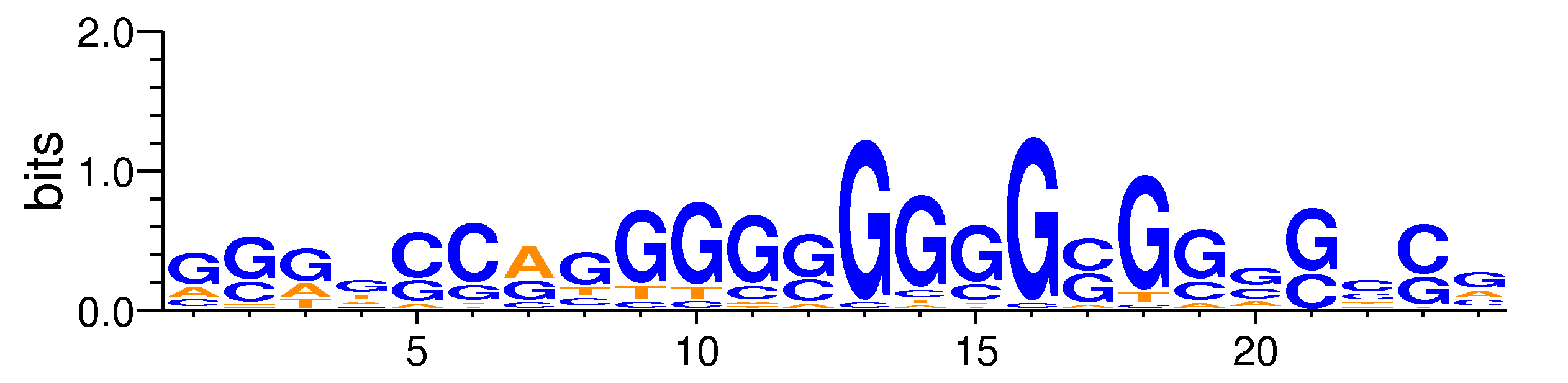
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SP1#MA0079.2 | -6 | 1.03031e-05 | 0.0049043 | 0.00959545 | 10 | GGGGCCAGGGGGGGGGCGGGGCCG | GGGGGCGGGG | - |
| opa#MA0456.1 | -5 | 7.49923e-05 | 0.0356963 | 0.0349207 | 12 | GGGGCCAGGGGGGGGGCGGGGCCG | CAGCGGGGGGTC | - |
| Klf4#MA0039.2 | -7 | 0.000323414 | 0.153945 | 0.0632492 | 10 | GGGGCCAGGGGGGGGGCGGGGCCG | TGGGTGGGGC | + |
| YGR067C#MA0425.1 | -3 | 0.000403767 | 0.192193 | 0.0632492 | 14 | GGGGCCAGGGGGGGGGCGGGGCCG | TGAAAAAGTGGGGT | - |
| SP1#MA0079.2 | -7 | 0.000471498 | 0.224433 | 0.0632492 | 10 | GGGGCCAGGGGGGGGGCGGGGCCG | GGGGCGGGGT | + |
| Pax4#MA0068.1 | 0 | 0.000498132 | 0.237111 | 0.0632492 | 24 | GGGGCCAGGGGGGGGGCGGGGCCG | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| btd#MA0443.1 | -10 | 0.000533242 | 0.253823 | 0.0632492 | 10 | GGGGCCAGGGGGGGGGCGGGGCCG | AGGGGGCGGA | + |
| MIG2#MA0338.1 | -9 | 0.000599782 | 0.285496 | 0.0632492 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | TGCGGGG | - |
| PLAG1#MA0163.1 | 0 | 0.000611226 | 0.290944 | 0.0632492 | 14 | GGGGCCAGGGGGGGGGCGGGGCCG | GGGGCCCAAGGGGG | + |
| abi4#MA0123.1 | -8 | 0.000861915 | 0.410272 | 0.0802713 | 10 | GGGGCCAGGGGGGGGGCGGGGCCG | GGGGGCACCG | - |
| RREB1#MA0073.1 | 0 | 0.00139622 | 0.6646 | 0.110909 | 20 | GGGGCCAGGGGGGGGGCGGGGCCG | TGGGGGGGGGTGGTTTGGGG | - |
| MIG3#MA0339.1 | -9 | 0.00142907 | 0.680237 | 0.110909 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | TGCGGGG | - |
| Pax5#MA0014.1 | -1 | 0.00164805 | 0.784471 | 0.118065 | 20 | GGGGCCAGGGGGGGGGCGGGGCCG | GGAGCACTGAAGCGTAACCG | + |
| STP2#MA0395.1 | -4 | 0.00228409 | 1.08723 | 0.151943 | 20 | GGGGCCAGGGGGGGGGCGGGGCCG | TGATCGGCGCCGCACGACGA | + |
| CTCF#MA0139.1 | -1 | 0.00278077 | 1.32365 | 0.172651 | 19 | GGGGCCAGGGGGGGGGCGGGGCCG | TGGCCACCAGGGGGCGCTA | + |
| Macho-1#MA0118.1 | -7 | 0.00319326 | 1.51999 | 0.174897 | 9 | GGGGCCAGGGGGGGGGCGGGGCCG | TGGGGGGTC | + |
| MSN2#MA0341.1 | -11 | 0.00350891 | 1.67024 | 0.174897 | 5 | GGGGCCAGGGGGGGGGCGGGGCCG | AGGGG | + |
| YML081W#MA0431.1 | -8 | 0.00356813 | 1.69843 | 0.174897 | 9 | GGGGCCAGGGGGGGGGCGGGGCCG | GTGCGGGGT | - |
| MIG1#MA0337.1 | -9 | 0.00421342 | 2.00559 | 0.196201 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | GCGGGGG | - |
| SUT1#MA0399.1 | -9 | 0.00448883 | 2.13668 | 0.199072 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | CGCGGGG | + |
| Zfx#MA0146.1 | -5 | 0.00470431 | 2.23925 | 0.199145 | 14 | GGGGCCAGGGGGGGGGCGGGGCCG | CAGGCCTCGGCCCC | - |
| IXR1#MA0323.1 | -1 | 0.00651377 | 3.10056 | 0.263755 | 15 | GGGGCCAGGGGGGGGGCGGGGCCG | AAGCCGGAAGCGGGG | + |
| DOT6#MA0351.1 | -5 | 0.00798208 | 3.79947 | 0.300995 | 19 | GGGGCCAGGGGGGGGGCGGGGCCG | AGGATGCGATGAGGTGCAGAA | - |
| Egr1#MA0162.1 | -8 | 0.00834212 | 3.97085 | 0.300995 | 11 | GGGGCCAGGGGGGGGGCGGGGCCG | TGCGTGGGCGT | + |
| YPR022C#MA0436.1 | -7 | 0.0087566 | 4.16814 | 0.300995 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | CGTGGGG | - |
| TFAP2A#MA0003.1 | -3 | 0.00925574 | 4.40573 | 0.300995 | 9 | GGGGCCAGGGGGGGGGCGGGGCCG | GCCCGGGGG | + |
| INSM1#MA0155.1 | -2 | 0.00949444 | 4.51935 | 0.300995 | 12 | GGGGCCAGGGGGGGGGCGGGGCCG | TGTCAGGGGGCG | + |
| UGA3#MA0410.1 | -8 | 0.00962812 | 4.58299 | 0.300995 | 8 | GGGGCCAGGGGGGGGGCGGGGCCG | CGGCGGGA | + |
| CRZ1#MA0285.1 | -12 | 0.00973623 | 4.63444 | 0.300995 | 9 | GGGGCCAGGGGGGGGGCGGGGCCG | GTGGCTTAG | - |
| ADR1#MA0268.1 | -8 | 0.0104232 | 4.96147 | 0.300995 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | GTGGGGT | - |
| RPN4#MA0373.1 | -11 | 0.0104232 | 4.96147 | 0.300995 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | GGTGGCG | + |
| MSN4#MA0342.1 | -6 | 0.0106654 | 5.07674 | 0.300995 | 5 | GGGGCCAGGGGGGGGGCGGGGCCG | AGGGG | + |
| MZF1_5-13#MA0057.1 | -7 | 0.0132641 | 6.31371 | 0.360371 | 10 | GGGGCCAGGGGGGGGGCGGGGCCG | GGAGGGGGAA | + |
| ESR1#MA0112.2 | -1 | 0.0135432 | 6.44658 | 0.360371 | 20 | GGGGCCAGGGGGGGGGCGGGGCCG | AGGTCAGGGTGACCTGGGCC | - |
| RGM1#MA0366.1 | -6 | 0.0142019 | 6.76012 | 0.367401 | 5 | GGGGCCAGGGGGGGGGCGGGGCCG | AGGGG | + |
| RSC30#MA0375.1 | -12 | 0.0162596 | 7.73958 | 0.384022 | 8 | GGGGCCAGGGGGGGGGCGGGGCCG | CGCGCGCG | - |
| REI1#MA0364.1 | -4 | 0.0163319 | 7.77397 | 0.384022 | 7 | GGGGCCAGGGGGGGGGCGGGGCCG | TCAGGGG | - |
| RXR::RAR_DR5#MA0159.1 | -1 | 0.0164938 | 7.85104 | 0.384022 | 17 | GGGGCCAGGGGGGGGGCGGGGCCG | AGGTCATGGAGAGGTCA | + |
| Mycn#MA0104.2 | -3 | 0.018301 | 8.71127 | 0.408299 | 10 | GGGGCCAGGGGGGGGGCGGGGCCG | GCCACGTGCG | - |
| MET31#MA0333.1 | -9 | 0.0184133 | 8.76473 | 0.408299 | 9 | GGGGCCAGGGGGGGGGCGGGGCCG | GGTGTGGCG | + |
| hkb#MA0450.1 | -7 | 0.0202403 | 9.63441 | 0.438374 | 9 | GGGGCCAGGGGGGGGGCGGGGCCG | GGGGCGTGA | + |