MacroAPE 1083:CNhs11828 bu f1
From FANTOM5_SSTAR
Full Name: CNhs11828_bu_f1
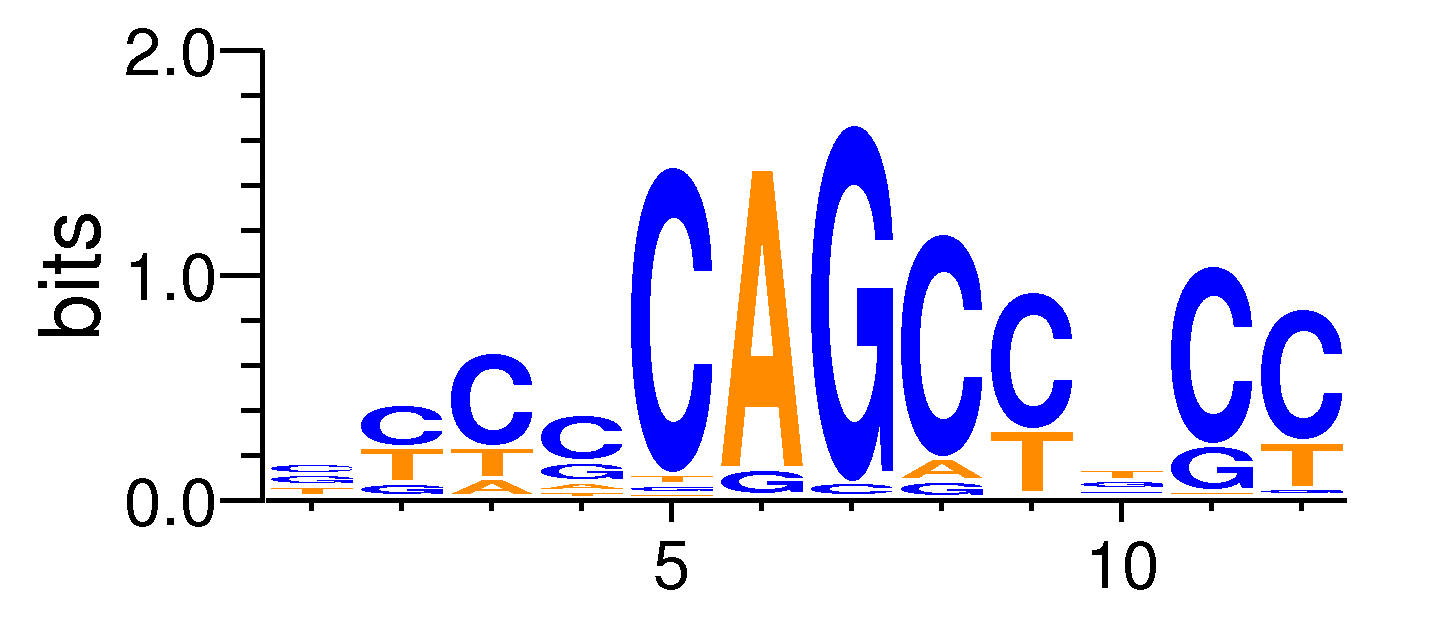
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SP1#MA0079.2 | -2 | 0.000240298 | 0.114382 | 0.127673 | 10 | CCCCCAGCCTCC | CCCCGCCCCC | + |
| CRZ1#MA0285.1 | -2 | 0.000268856 | 0.127975 | 0.127673 | 9 | CCCCCAGCCTCC | CTAAGCCAC | + |
| ACE2#MA0267.1 | -2 | 0.0014326 | 0.681917 | 0.453536 | 7 | CCCCCAGCCTCC | ACCAGCA | + |
| SP1#MA0079.2 | -1 | 0.00278302 | 1.32472 | 0.465559 | 10 | CCCCCAGCCTCC | ACCCCGCCCC | - |
| NHLH1#MA0048.1 | -1 | 0.002854 | 1.3585 | 0.465559 | 11 | CCCCCAGCCTCC | GCGCAGCTGCGT | + |
| abi4#MA0123.1 | -2 | 0.00294115 | 1.39999 | 0.465559 | 10 | CCCCCAGCCTCC | CGGTGCCCCC | + |
| Myf#MA0055.1 | -1 | 0.00507353 | 2.415 | 0.507521 | 11 | CCCCCAGCCTCC | CAGCAGCTGCTG | - |
| znf143#MA0088.1 | 3 | 0.00512305 | 2.43857 | 0.507521 | 12 | CCCCCAGCCTCC | GATTTCCCATAATGCCTTGC | + |
| IXR1#MA0323.1 | -2 | 0.00534374 | 2.54362 | 0.507521 | 10 | CCCCCAGCCTCC | CCCCGCTTCCGGCTT | - |
| SWI5#MA0402.1 | -1 | 0.00894509 | 4.25786 | 0.772327 | 8 | CCCCCAGCCTCC | AACCAGCA | - |
| Zfx#MA0146.1 | 3 | 0.0100253 | 4.77206 | 0.783019 | 11 | CCCCCAGCCTCC | CAGGCCTCGGCCCC | - |
| Pax4#MA0068.1 | 14 | 0.0107178 | 5.10168 | 0.783019 | 12 | CCCCCAGCCTCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| Myb#MA0100.1 | -4 | 0.0123475 | 5.87741 | 0.792922 | 8 | CCCCCAGCCTCC | CAACGGCC | - |
| Gamyb#MA0034.1 | -2 | 0.0125231 | 5.96101 | 0.792922 | 10 | CCCCCAGCCTCC | GACAACCGCC | + |
| YGR067C#MA0425.1 | 0 | 0.0168064 | 7.99984 | 0.8794 | 12 | CCCCCAGCCTCC | ACCCCACTTTTTTG | + |
| NHP10#MA0344.1 | 0 | 0.016854 | 8.02251 | 0.8794 | 8 | CCCCCAGCCTCC | TCCCCGGC | - |
| btd#MA0443.1 | -3 | 0.0174627 | 8.31223 | 0.8794 | 9 | CCCCCAGCCTCC | TCCGCCCCCT | - |
| CHA4#MA0283.1 | -1 | 0.0206163 | 9.81338 | 0.95028 | 8 | CCCCCAGCCTCC | TCTCCGCC | - |