MacroAPE 1083:CEBPB ThioMac-CEBPb-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: CEBPB_ThioMac-CEBPb-ChIP-Seq/Homer
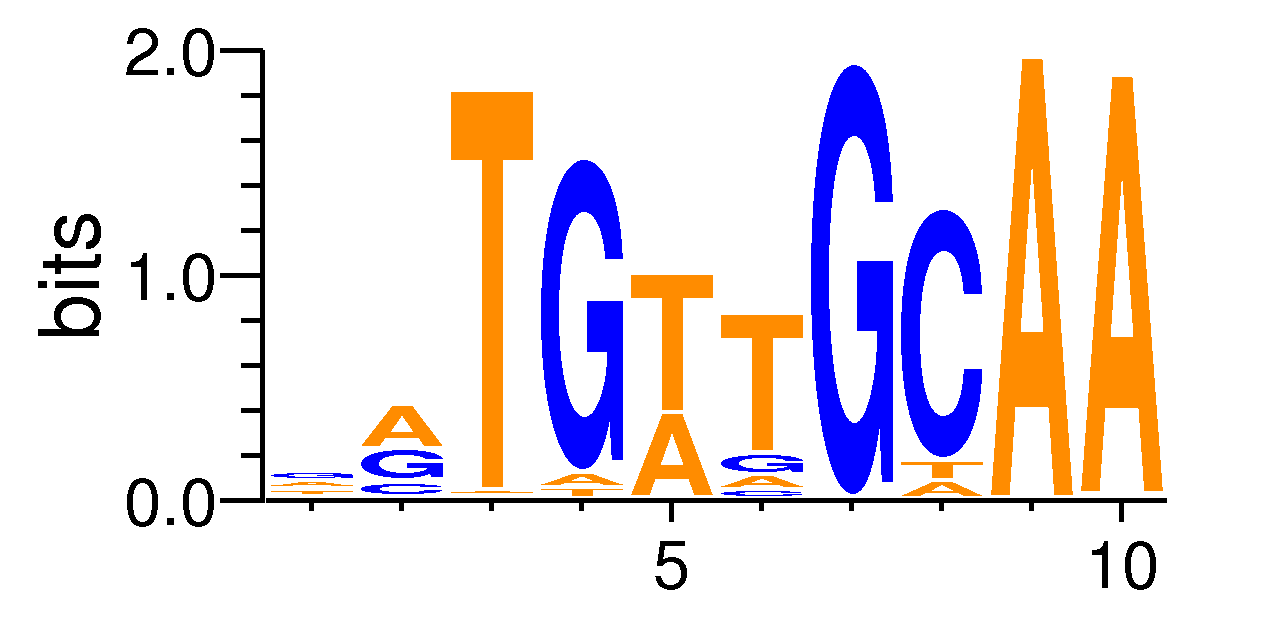
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| mab-3#MA0262.1 | 0 | 0.000761446 | 0.362448 | 0.724896 | 10 | GATGTTGCAA | AATGTTGCGAATT | + |
| CEBPA#MA0102.2 | -2 | 0.00221978 | 1.05661 | 0.861074 | 8 | GATGTTGCAA | TTTCGCAAT | + |
| Cebpa#MA0102.1 | 0 | 0.00501882 | 2.38896 | 0.861074 | 10 | GATGTTGCAA | GAGATTGCGAAA | - |
| Pou5f1#MA0142.1 | 3 | 0.00542216 | 2.58095 | 0.861074 | 10 | GATGTTGCAA | CTTTGTTATGCAAAT | + |
| RFX1#MA0365.1 | -2 | 0.00613427 | 2.91991 | 0.861074 | 8 | GATGTTGCAA | GGTTGCTA | + |
| STE12#MA0393.1 | -2 | 0.00666041 | 3.17036 | 0.861074 | 7 | GATGTTGCAA | TGTTTCA | - |
| Ddit3::Cebpa#MA0019.1 | -2 | 0.00745716 | 3.54961 | 0.861074 | 8 | GATGTTGCAA | AGATGCAATCCC | + |
| Sox2#MA0143.1 | 4 | 0.0081404 | 3.87483 | 0.861074 | 10 | GATGTTGCAA | CCTTTGTTATGCAAA | + |
| BRCA1#MA0133.1 | -1 | 0.0163287 | 7.77245 | 1 | 7 | GATGTTGCAA | GTGTTGT | - |
| slbo#MA0244.1 | -3 | 0.0202222 | 9.62575 | 1 | 7 | GATGTTGCAA | TTTGCAAT | - |
| HLF#MA0043.1 | 0 | 0.0204685 | 9.74299 | 1 | 10 | GATGTTGCAA | GGTTACGCAATT | + |