MacroAPE 1083:CNhs10752 bl f1
From FANTOM5_SSTAR
Full Name: CNhs10752_bl_f1
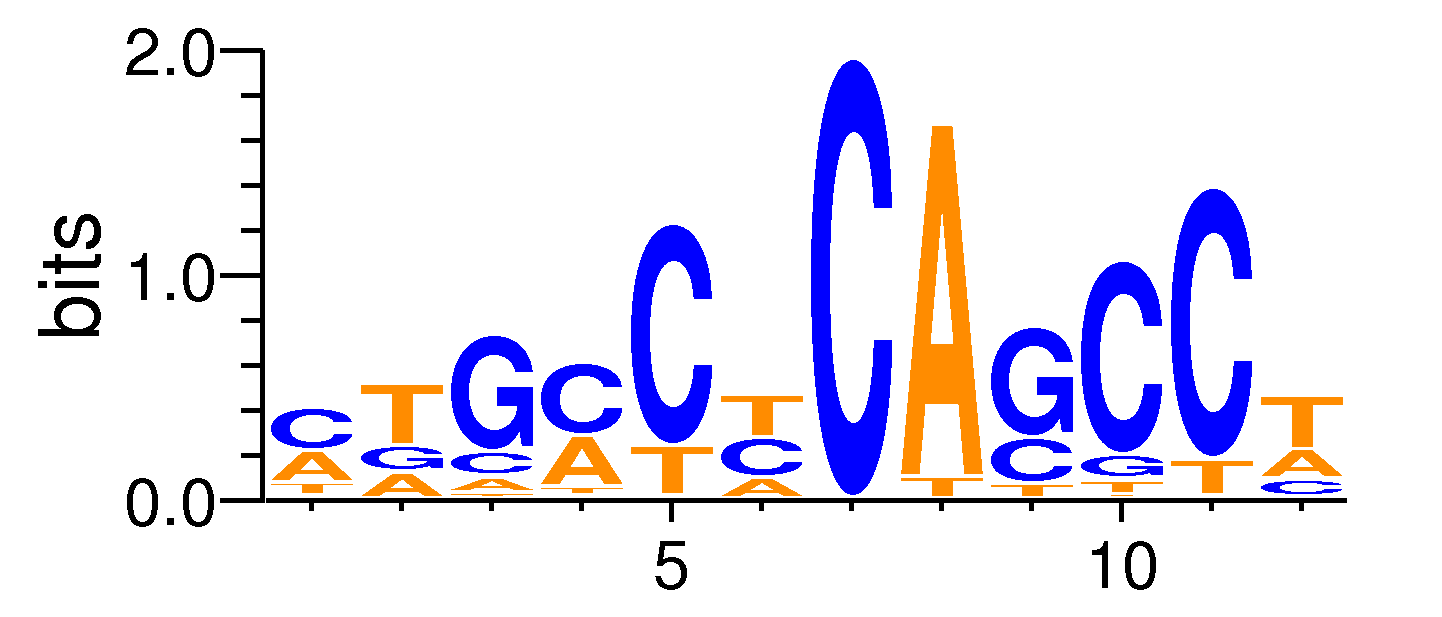
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Klf4#MA0039.2 | -2 | 6.6171e-05 | 0.0314974 | 0.0628291 | 10 | CTGCCTCAGCCT | GCCCCACCCA | - |
| NFE2L2#MA0150.1 | 0 | 0.00125777 | 0.598698 | 0.438314 | 11 | CTGCCTCAGCCT | ATGACTCAGCA | + |
| CRZ1#MA0285.1 | -4 | 0.00138489 | 0.659206 | 0.438314 | 8 | CTGCCTCAGCCT | CTAAGCCAC | + |
| Tcfcp2l1#MA0145.1 | 1 | 0.00189748 | 0.903202 | 0.450413 | 12 | CTGCCTCAGCCT | CCAGTTCAAACCAG | + |
| Mafb#MA0117.1 | -2 | 0.00408993 | 1.9468 | 0.653373 | 8 | CTGCCTCAGCCT | GCGTCAGC | - |
| Zfx#MA0146.1 | 1 | 0.00466662 | 2.22131 | 0.653373 | 12 | CTGCCTCAGCCT | GGGGCCGAGGCCTG | + |
| SP1#MA0079.1 | -3 | 0.00647333 | 3.0813 | 0.653373 | 9 | CTGCCTCAGCCT | ACCCCGCCCC | - |
| CHA4#MA0283.1 | -3 | 0.0081346 | 3.87207 | 0.653373 | 8 | CTGCCTCAGCCT | TCTCCGCC | - |
| Tal1::Gata1#MA0140.1 | 5 | 0.00818755 | 3.89727 | 0.653373 | 12 | CTGCCTCAGCCT | CTTATCTGTCCCCACCAG | - |
| AP1#MA0099.2 | -1 | 0.00824112 | 3.92277 | 0.653373 | 7 | CTGCCTCAGCCT | TGACTCA | + |
| Pax4#MA0068.1 | 12 | 0.0088335 | 4.20475 | 0.653373 | 12 | CTGCCTCAGCCT | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| GCN4#MA0303.1 | 6 | 0.00974184 | 4.63712 | 0.653373 | 12 | CTGCCTCAGCCT | TGAAGTATGACTCATCCCTTG | - |
| RUNX1#MA0002.2 | 0 | 0.0107521 | 5.11799 | 0.653373 | 11 | CTGCCTCAGCCT | AAACCACAGAC | - |
| AFT2#MA0270.1 | -4 | 0.0115652 | 5.50505 | 0.653373 | 8 | CTGCCTCAGCCT | CACACCCC | + |
| TFAP2A#MA0003.1 | -2 | 0.0119931 | 5.70872 | 0.653373 | 9 | CTGCCTCAGCCT | CCCCCGGGC | - |
| ACE2#MA0267.1 | -4 | 0.0125584 | 5.97778 | 0.653373 | 7 | CTGCCTCAGCCT | ACCAGCA | + |
| BAS1#MA0278.1 | 5 | 0.0126398 | 6.01653 | 0.653373 | 12 | CTGCCTCAGCCT | TTGACTTGACTCTGGCTGTGC | - |