MacroAPE 1083:FOXA1 LNCAP-FOXA1-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: FOXA1_LNCAP-FOXA1-ChIP-Seq/Homer
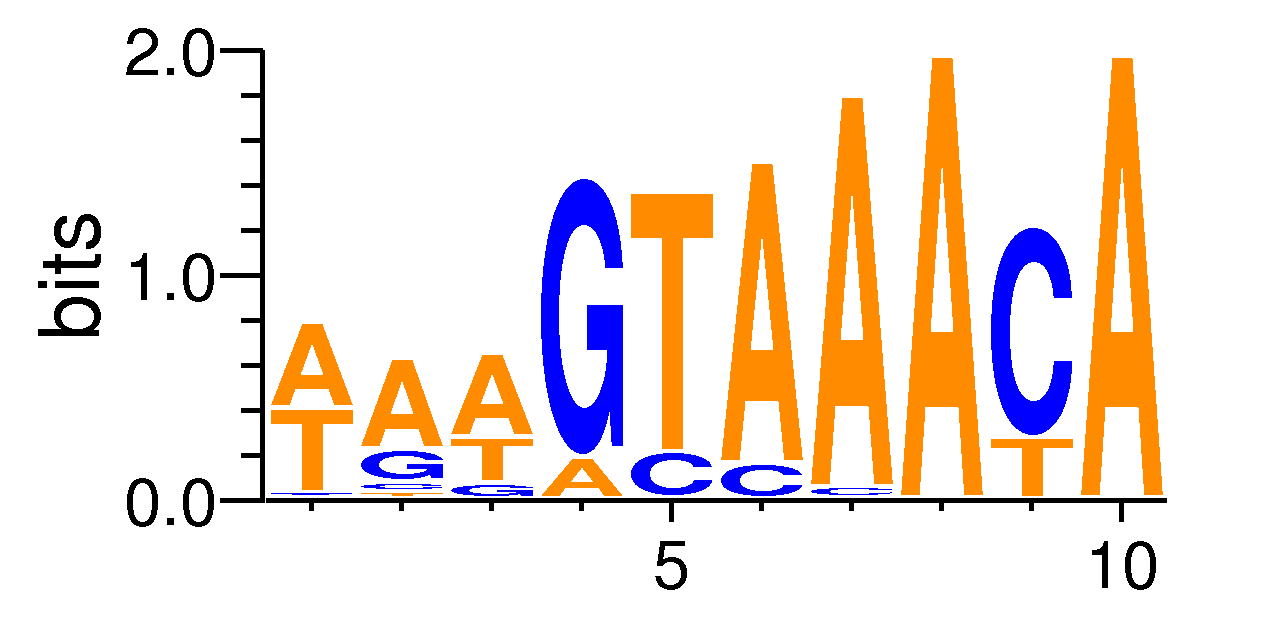
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Foxa2#MA0047.2 | 2 | 1.57604e-12 | 7.50196e-10 | 1.481e-09 | 10 | AAAGTAAACA | CCTAAGTAAACA | - |
| FOXA1#MA0148.1 | 1 | 2.74075e-09 | 1.3046e-06 | 1.28774e-06 | 10 | AAAGTAAACA | CAAAGTAAACA | - |
| FKH2#MA0297.1 | -3 | 1.23738e-06 | 0.000588993 | 0.000387588 | 7 | AAAGTAAACA | GTAAACA | + |
| fkh#MA0446.1 | 1 | 4.36303e-06 | 0.0020768 | 0.00102498 | 10 | AAAGTAAACA | TTAAGCAAACA | - |
| slp1#MA0458.1 | 0 | 1.05031e-05 | 0.00499949 | 0.00197395 | 10 | AAAGTAAACA | AATGTAAACAA | - |
| FKH1#MA0296.1 | 3 | 2.5415e-05 | 0.0120976 | 0.00394188 | 10 | AAAGTAAACA | TGAATTGTAAACAAAGAGGA | + |
| FOXD1#MA0031.1 | -3 | 2.93639e-05 | 0.0139772 | 0.00394188 | 7 | AAAGTAAACA | GTAAACAT | + |
| FOXO3#MA0157.1 | -2 | 5.31832e-05 | 0.0253152 | 0.00624702 | 8 | AAAGTAAACA | TGTAAACA | + |
| Foxa2#MA0047.2 | -1 | 6.21599e-05 | 0.0295881 | 0.00627884 | 9 | AAAGTAAACA | AAGTAAATATTG | - |
| FOXF2#MA0030.1 | 2 | 6.68176e-05 | 0.0318052 | 0.00627884 | 10 | AAAGTAAACA | CAAACGTAAACAAT | + |
| br_Z4#MA0013.1 | -1 | 0.000247412 | 0.117768 | 0.0211357 | 9 | AAAGTAAACA | TAGTAAACAAA | + |
| Foxd3#MA0041.1 | -1 | 0.000483877 | 0.230326 | 0.0378916 | 9 | AAAGTAAACA | AAACAAACATTC | - |
| FOXI1#MA0042.1 | -1 | 0.000555173 | 0.264262 | 0.0401304 | 9 | AAAGTAAACA | AAACAAACATCC | - |
| HCM1#MA0317.1 | -3 | 0.000661718 | 0.314978 | 0.0444154 | 7 | AAAGTAAACA | ATAAACAA | + |
| Foxq1#MA0040.1 | -2 | 0.00145839 | 0.694192 | 0.0913629 | 8 | AAAGTAAACA | AATAAACAATA | - |
| SRY#MA0084.1 | -3 | 0.00218482 | 1.03998 | 0.128317 | 7 | AAAGTAAACA | GTAAACAAT | + |
| mirr#MA0233.1 | -5 | 0.00423152 | 2.0142 | 0.233903 | 5 | AAAGTAAACA | AAACA | + |
| ara#MA0210.1 | -5 | 0.00639859 | 3.04573 | 0.334042 | 5 | AAAGTAAACA | TAACA | + |
| YAP7#MA0419.1 | 1 | 0.013576 | 6.46219 | 0.654008 | 10 | AAAGTAAACA | ATTAGTAAGCA | + |
| HMG-I/Y#MA0045.1 | 5 | 0.0139195 | 6.62569 | 0.654008 | 10 | AAAGTAAACA | CAACAAATGGAAAAAC | + |
| ELF5#MA0136.1 | -1 | 0.019024 | 9.05543 | 0.828698 | 9 | AAAGTAAACA | AAGGAAGTA | - |
| CG11617#MA0173.1 | -1 | 0.0199761 | 9.50862 | 0.828698 | 7 | AAAGTAAACA | ATGTTAA | - |
| caup#MA0217.1 | -5 | 0.0202832 | 9.65478 | 0.828698 | 5 | AAAGTAAACA | TAACA | + |