MacroAPE 1083:AR LNCAP-AR-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: AR_LNCAP-AR-ChIP-Seq/Homer
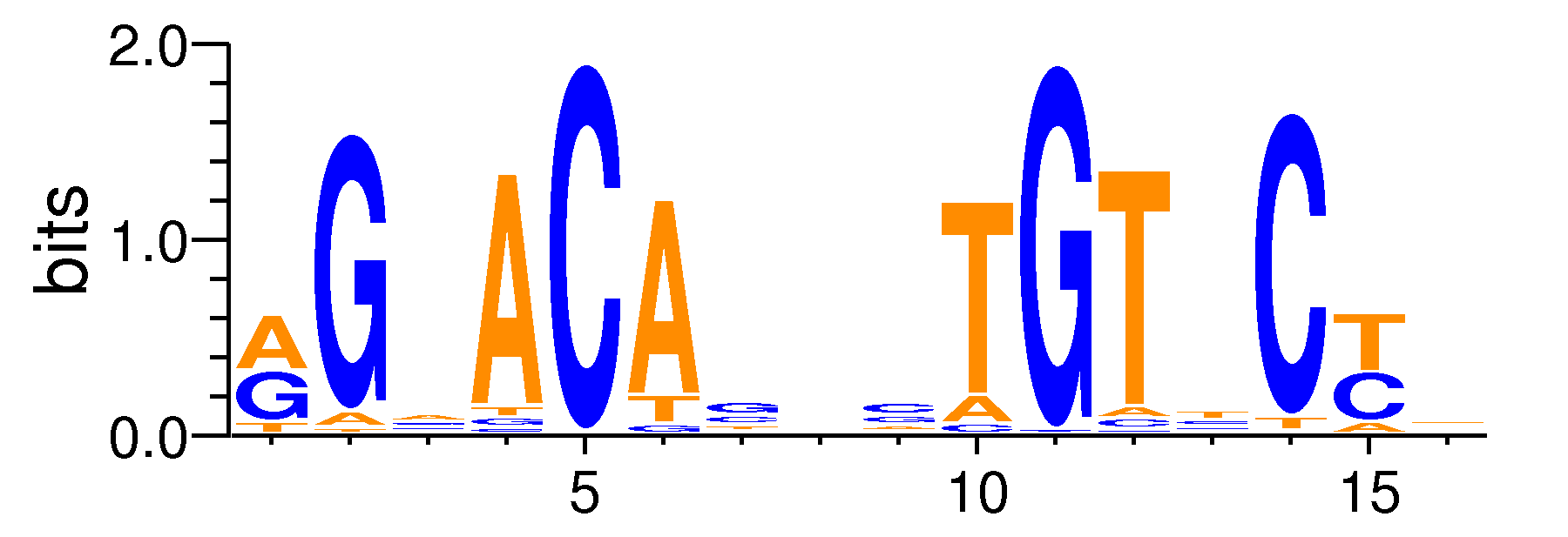
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Ar#MA0007.1 | 3 | 1.0814e-09 | 5.14745e-07 | 5.08093e-07 | 16 | AGAACAGTCTGTTCTT | ATAAGAACACCCTGTACCCGCC | + |
| NR3C1#MA0113.1 | 2 | 2.32895e-08 | 1.10858e-05 | 7.29503e-06 | 16 | AGAACAGTCTGTTCTT | TCAGGACATAATGTTCCC | - |
| ESR2#MA0258.1 | 2 | 3.39298e-05 | 0.0161506 | 0.00611152 | 16 | AGAACAGTCTGTTCTT | CAAGGTCACGGTGACCTG | + |
| ESR1#MA0112.2 | 0 | 0.000284945 | 0.135634 | 0.0382518 | 16 | AGAACAGTCTGTTCTT | AGGTCAGGGTGACCTGGGCC | - |
| GAL4#MA0299.1 | 1 | 0.00156576 | 0.7453 | 0.154955 | 14 | AGAACAGTCTGTTCTT | CGGGGAACAGTACTC | + |
| lin-14#MA0261.1 | -8 | 0.00164899 | 0.784918 | 0.154955 | 6 | AGAACAGTCTGTTCTT | GTGTTC | - |
| MGA1#MA0336.1 | -2 | 0.0031471 | 1.49802 | 0.232505 | 14 | AGAACAGTCTGTTCTT | TTTAGAGTGTTCTATAGTCCT | - |
| ESR1#MA0112.2 | 1 | 0.00361425 | 1.72038 | 0.242593 | 16 | AGAACAGTCTGTTCTT | GGGGTCACGGTGACCTGG | - |
| PPARG#MA0066.1 | 2 | 0.00466621 | 2.22111 | 0.260065 | 16 | AGAACAGTCTGTTCTT | GTAGGTCACGGTGACCTACT | + |
| RXRA::VDR#MA0074.1 | 0 | 0.0138129 | 6.57493 | 0.683154 | 15 | AGAACAGTCTGTTCTT | GGGTCAACGAGTTCA | + |