MacroAPE 1083:CNhs13801 bl f2
From FANTOM5_SSTAR
Full Name: CNhs13801_bl_f2
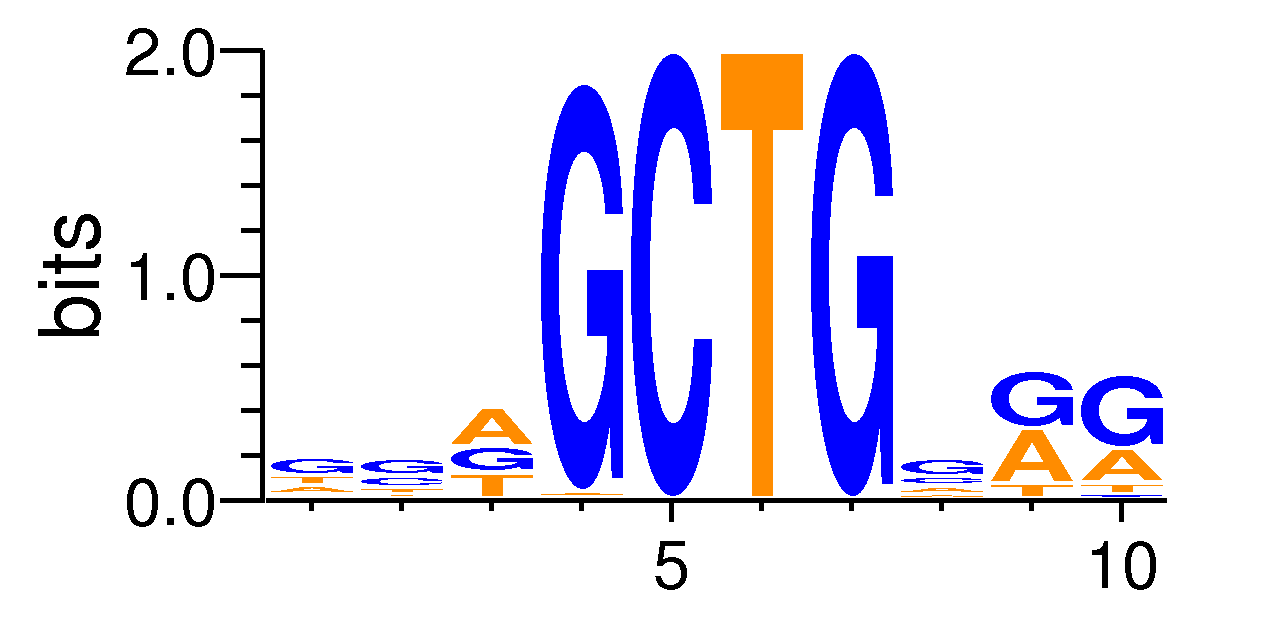
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| NHLH1#MA0048.1 | 2 | 0.00130149 | 0.619508 | 0.482762 | 10 | GGAGCTGGGG | ACGCAGCTGCGC | - |
| ACE2#MA0267.1 | -2 | 0.00203679 | 0.969513 | 0.482762 | 7 | GGAGCTGGGG | TGCTGGT | - |
| Tcfcp2l1#MA0145.1 | 6 | 0.00233157 | 1.10983 | 0.482762 | 8 | GGAGCTGGGG | CTGGTTTGAACTGG | - |
| Myf#MA0055.1 | 2 | 0.00253884 | 1.20849 | 0.482762 | 10 | GGAGCTGGGG | CAGCAGCTGCTG | + |
| CRZ1#MA0285.1 | 0 | 0.00464958 | 2.2132 | 0.736768 | 9 | GGAGCTGGGG | GTGGCTTAG | - |
| SWI5#MA0402.1 | -2 | 0.00612134 | 2.91376 | 0.831413 | 8 | GGAGCTGGGG | TGCTGGTT | + |
| Zfx#MA0146.1 | 0 | 0.0101047 | 4.80982 | 0.960299 | 10 | GGAGCTGGGG | GGGGCCGAGGCCTG | + |
| REST#MA0138.2 | 11 | 0.010997 | 5.23455 | 0.960299 | 8 | GGAGCTGGGG | GCGCTGTCCATGGTGCTGA | + |
| SP1#MA0079.1 | 0 | 0.011227 | 5.34405 | 0.960299 | 10 | GGAGCTGGGG | GGGGCGGGGT | + |
| BRCA1#MA0133.1 | -1 | 0.0116812 | 5.56024 | 0.960299 | 7 | GGAGCTGGGG | GTGTTGT | - |
| Mafb#MA0117.1 | -3 | 0.0121205 | 5.76935 | 0.960299 | 7 | GGAGCTGGGG | GCTGACGG | + |
| REST#MA0138.2 | 12 | 0.014466 | 6.88582 | 0.998691 | 9 | GGAGCTGGGG | GGCGCTGTCCATGGTGCTGAA | - |
| Pax4#MA0068.1 | 5 | 0.0172816 | 8.22604 | 0.998691 | 10 | GGAGCTGGGG | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |