MacroAPE 1083:THB do
From FANTOM5_SSTAR
Full Name: THB_do
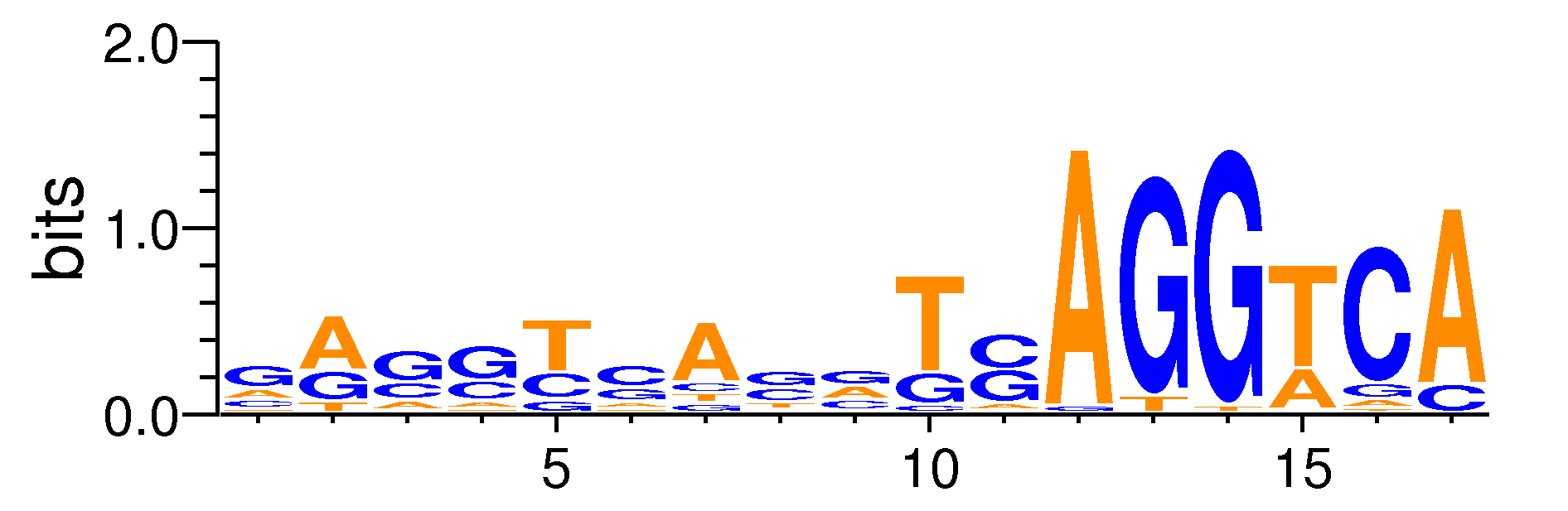
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| NR4A2#MA0160.1 | -10 | 0.000666921 | 0.317454 | 0.251106 | 7 | GAGGTCAGGTCAGGTCA | AAGGTCAC | + |
| CREB1#MA0018.2 | -9 | 0.000715795 | 0.340719 | 0.251106 | 8 | GAGGTCAGGTCAGGTCA | TGACGTCA | + |
| RXR::RAR_DR5#MA0159.1 | 0 | 0.000845508 | 0.402462 | 0.251106 | 17 | GAGGTCAGGTCAGGTCA | AGGTCATGGAGAGGTCA | + |
| Tal1::Gata1#MA0140.1 | 0 | 0.00122132 | 0.58135 | 0.251106 | 17 | GAGGTCAGGTCAGGTCA | CTGGTGGGGACAGATAAG | + |
| Esrrb#MA0141.1 | -5 | 0.00171318 | 0.815472 | 0.251106 | 12 | GAGGTCAGGTCAGGTCA | AGCTCAAGGTCA | + |
| ESR1#MA0112.2 | -6 | 0.00188708 | 0.898252 | 0.251106 | 11 | GAGGTCAGGTCAGGTCA | GGCCCAGGTCACCCTGACCT | + |
| DOT6#MA0351.1 | 0 | 0.00211962 | 1.00894 | 0.251106 | 17 | GAGGTCAGGTCAGGTCA | AGGATGCGATGAGGTGCAGAA | - |
| YAP6#MA0418.1 | -2 | 0.00248704 | 1.18383 | 0.261896 | 15 | GAGGTCAGGTCAGGTCA | GGTCTGATTACGTAAGCGAC | + |
| TOD6#MA0350.1 | 0 | 0.00699136 | 3.32789 | 0.601305 | 17 | GAGGTCAGGTCAGGTCA | AAAACGCGATGAGCTGTGCCT | - |
| CST6#MA0286.1 | -8 | 0.00761355 | 3.62405 | 0.601305 | 9 | GAGGTCAGGTCAGGTCA | ATGACGTAA | + |
| CREB1#MA0018.2 | -4 | 0.00945551 | 4.50082 | 0.652273 | 12 | GAGGTCAGGTCAGGTCA | CCTGGTGACGTC | + |
| ASH1#MA0276.1 | -4 | 0.0102346 | 4.87165 | 0.652273 | 10 | GAGGTCAGGTCAGGTCA | CCGGATCGGG | + |
| YAP1#MA0415.1 | 2 | 0.0103236 | 4.91404 | 0.652273 | 17 | GAGGTCAGGTCAGGTCA | ACGAGCTTACGTAAGCAAAT | - |
| bZIP910#MA0096.1 | -11 | 0.0123585 | 5.88265 | 0.683078 | 6 | GAGGTCAGGTCAGGTCA | ACGTCAT | - |
| RORA_1#MA0071.1 | -7 | 0.0125091 | 5.95433 | 0.683078 | 10 | GAGGTCAGGTCAGGTCA | ATCAAGGTCA | + |
| PPARG::RXRA#MA0065.2 | -2 | 0.0129734 | 6.17534 | 0.683078 | 15 | GAGGTCAGGTCAGGTCA | GTAGGGCAAAGGTCA | + |
| Pax4#MA0068.1 | 6 | 0.014007 | 6.66733 | 0.698682 | 17 | GAGGTCAGGTCAGGTCA | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| BAS1#MA0278.1 | 3 | 0.0151995 | 7.23495 | 0.706768 | 17 | GAGGTCAGGTCAGGTCA | GCACAGCCAGAGTCAAGTCAA | + |
| RORA_2#MA0072.1 | -4 | 0.0157127 | 7.47925 | 0.706768 | 13 | GAGGTCAGGTCAGGTCA | TATAAGTAGGTCAA | + |
| YGR067C#MA0425.1 | -1 | 0.0164063 | 7.80941 | 0.706768 | 14 | GAGGTCAGGTCAGGTCA | TGAAAAAGTGGGGT | - |
| TGA1A#MA0129.1 | -10 | 0.0199042 | 9.4744 | 0.777333 | 7 | GAGGTCAGGTCAGGTCA | TACGTCA | + |