MacroAPE 1083:CNhs12525 bl f0
From FANTOM5_SSTAR
Full Name: CNhs12525_bl_f0
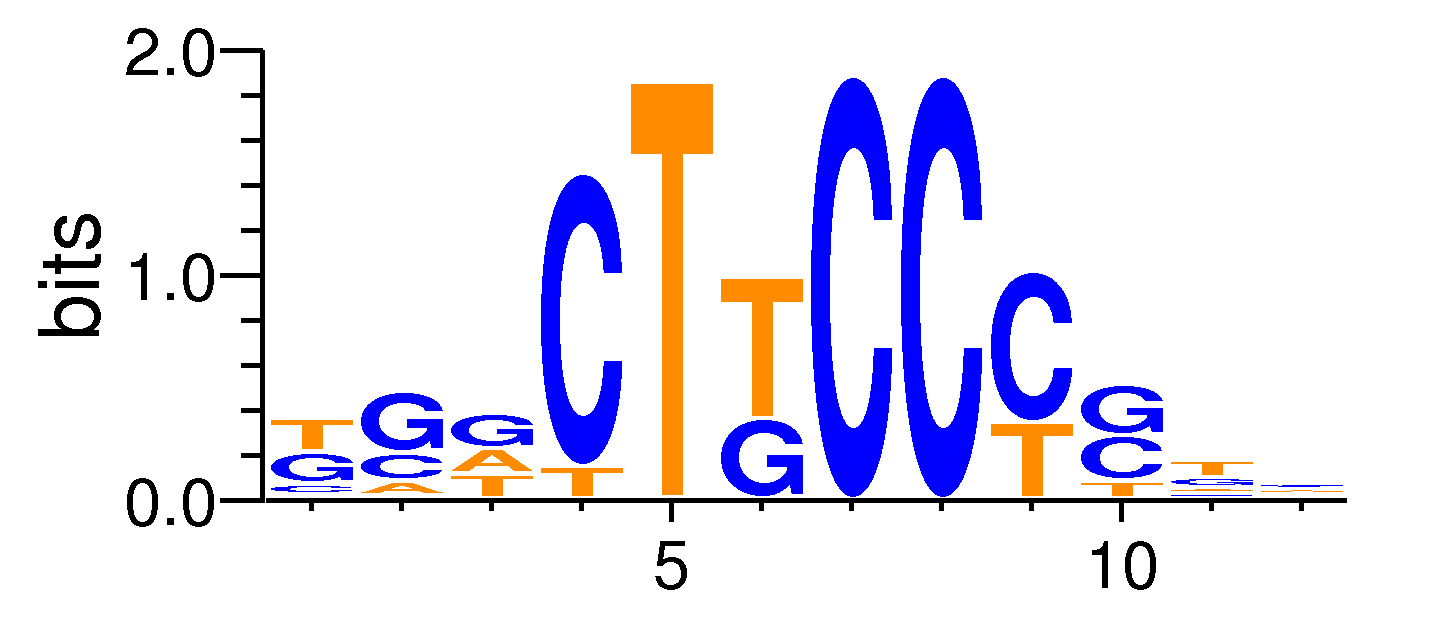
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| GCR2#MA0305.1 | -2 | 0.000113795 | 0.0541665 | 0.0781347 | 7 | TGGCTTCCCGTG | GCTTCCT | + |
| GABPA#MA0062.2 | -1 | 0.000241825 | 0.115109 | 0.0781347 | 10 | TGGCTTCCCGTG | CTCTTCCGGT | - |
| SPI1#MA0080.2 | -3 | 0.000274999 | 0.130899 | 0.0781347 | 6 | TGGCTTCCCGTG | CTTCCG | - |
| GCR1#MA0304.1 | -1 | 0.000331531 | 0.157809 | 0.0781347 | 8 | TGGCTTCCCGTG | GGCTTCCA | - |
| SPI1#MA0080.2 | -2 | 0.000808347 | 0.384773 | 0.152408 | 7 | TGGCTTCCCGTG | ACTTCCT | - |
| NF-kappaB#MA0061.1 | 1 | 0.00209139 | 0.995501 | 0.328118 | 9 | TGGCTTCCCGTG | GGGAATTTCC | + |
| GABPA#MA0062.2 | 1 | 0.0024364 | 1.15973 | 0.328118 | 10 | TGGCTTCCCGTG | GCCACTTCCGG | - |
| Eip74EF#MA0026.1 | -3 | 0.00323368 | 1.53923 | 0.335672 | 7 | TGGCTTCCCGTG | CTTCCGG | - |
| Stat3#MA0144.1 | -3 | 0.00365918 | 1.74177 | 0.335672 | 9 | TGGCTTCCCGTG | CTTCCTGGAA | - |
| ELK4#MA0076.1 | -2 | 0.00392062 | 1.86621 | 0.335672 | 9 | TGGCTTCCCGTG | ACTTCCGGT | - |
| ELK1#MA0028.1 | -3 | 0.00412554 | 1.96376 | 0.335672 | 9 | TGGCTTCCCGTG | CTTCCGGCTC | - |
| SPIB#MA0081.1 | -4 | 0.00472728 | 2.25019 | 0.335672 | 7 | TGGCTTCCCGTG | TTCCTCT | - |
| STAT1#MA0137.2 | -1 | 0.0048451 | 2.30627 | 0.335672 | 11 | TGGCTTCCCGTG | CATTTCCCGGAAACC | + |
| FEV#MA0156.1 | -2 | 0.00498498 | 2.37285 | 0.335672 | 8 | TGGCTTCCCGTG | ATTTCCTG | - |
| ELF5#MA0136.1 | -1 | 0.00593955 | 2.82723 | 0.373287 | 9 | TGGCTTCCCGTG | TACTTCCTT | + |
| che-1#MA0260.1 | -1 | 0.00646529 | 3.07748 | 0.380932 | 6 | TGGCTTCCCGTG | GGTTTC | - |
| NFKB1#MA0105.1 | 1 | 0.00707029 | 3.36546 | 0.392074 | 10 | TGGCTTCCCGTG | GGGGATTCCCC | + |
| ETS1#MA0098.1 | -3 | 0.00781364 | 3.71929 | 0.409223 | 6 | TGGCTTCCCGTG | CTTCCG | + |
| Klf4#MA0039.1 | -2 | 0.00918238 | 4.37081 | 0.455598 | 10 | TGGCTTCCCGTG | CCTTCCTTTA | - |
| REL#MA0101.1 | 2 | 0.0133805 | 6.3691 | 0.630697 | 8 | TGGCTTCCCGTG | GGGGATTTCC | + |
| dl_2#MA0023.1 | 2 | 0.0164811 | 7.84502 | 0.662001 | 8 | TGGCTTCCCGTG | GGGGATTTCC | + |
| znf143#MA0088.1 | -1 | 0.0169974 | 8.09078 | 0.662001 | 11 | TGGCTTCCCGTG | GATTTCCCATAATGCCTTGC | + |
| PUT3#MA0358.1 | -4 | 0.017223 | 8.19815 | 0.662001 | 8 | TGGCTTCCCGTG | TTCCCGGG | - |
| abi4#MA0123.1 | -1 | 0.018258 | 8.69079 | 0.662001 | 10 | TGGCTTCCCGTG | CGGTGCCCCC | + |
| PHD1#MA0355.1 | -1 | 0.018258 | 8.69079 | 0.662001 | 10 | TGGCTTCCCGTG | TGCTGCAGGT | - |
| ARO80#MA0273.1 | 2 | 0.0202711 | 9.64906 | 0.707773 | 12 | TGGCTTCCCGTG | ACATACTTGCCGAGAATTATC | - |