MacroAPE 1083:CNhs10634 bl f2
From FANTOM5_SSTAR
Full Name: CNhs10634_bl_f2
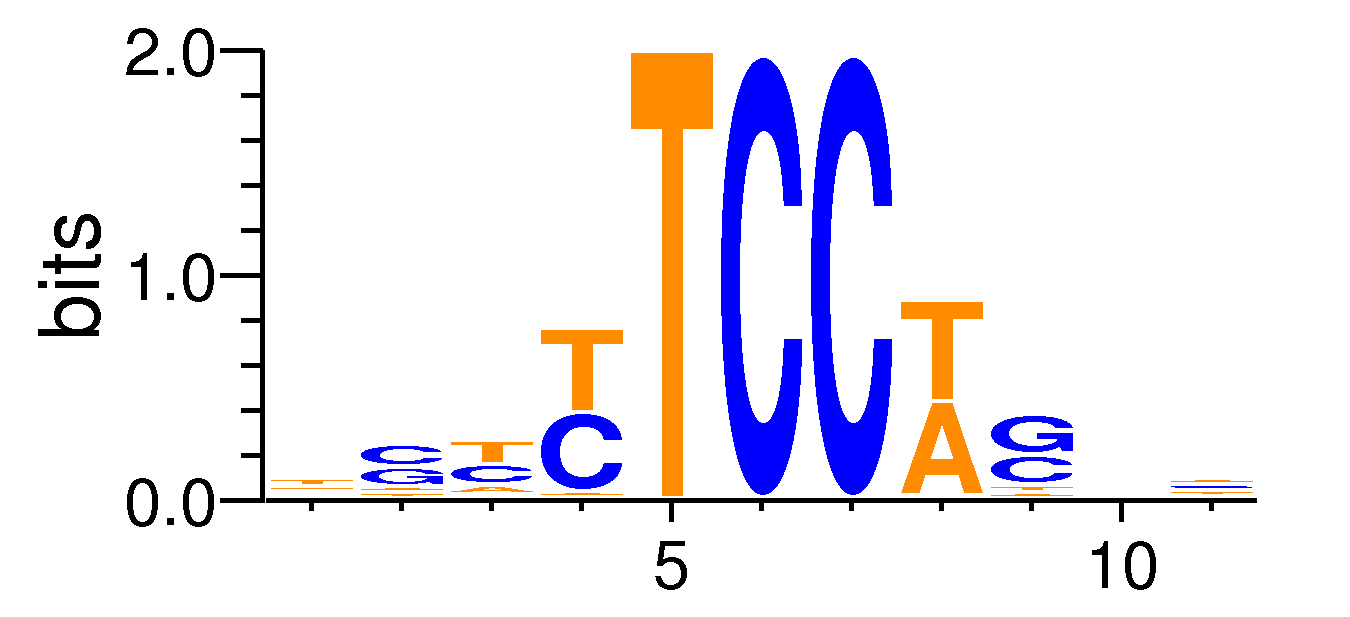
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| ELK1#MA0028.1 | -2 | 0.0017568 | 0.836235 | 0.646982 | 9 | TCTTTCCTGCA | CTTCCGGCTC | - |
| ELF5#MA0136.1 | 0 | 0.00212608 | 1.01201 | 0.646982 | 9 | TCTTTCCTGCA | TACTTCCTT | + |
| GCR2#MA0305.1 | -1 | 0.00328935 | 1.56573 | 0.646982 | 7 | TCTTTCCTGCA | GCTTCCT | + |
| Stat3#MA0144.1 | -2 | 0.00392541 | 1.8685 | 0.646982 | 9 | TCTTTCCTGCA | CTTCCTGGAA | - |
| SPIB#MA0081.1 | -3 | 0.00468604 | 2.23056 | 0.646982 | 7 | TCTTTCCTGCA | TTCCTCT | - |
| ETS1#MA0098.1 | -2 | 0.00477514 | 2.27297 | 0.646982 | 6 | TCTTTCCTGCA | CTTCCG | + |
| Klf4#MA0039.1 | -1 | 0.0054402 | 2.58954 | 0.646982 | 10 | TCTTTCCTGCA | CCTTCCTTTA | - |
| REL#MA0101.1 | 3 | 0.00686816 | 3.26924 | 0.646982 | 7 | TCTTTCCTGCA | GGGGATTTCC | + |
| TEAD1#MA0090.1 | 1 | 0.00708669 | 3.37326 | 0.646982 | 11 | TCTTTCCTGCA | CACATTCCTCCG | + |
| STAT1#MA0137.2 | 0 | 0.0075767 | 3.60651 | 0.646982 | 11 | TCTTTCCTGCA | GGTTTCCGGGAAATG | - |
| Eip74EF#MA0026.1 | -2 | 0.00814564 | 3.87733 | 0.646982 | 7 | TCTTTCCTGCA | CTTCCGG | - |
| FEV#MA0156.1 | -1 | 0.0114614 | 5.45564 | 0.838231 | 8 | TCTTTCCTGCA | ATTTCCTG | - |
| Tal1::Gata1#MA0140.1 | 6 | 0.0142125 | 6.76513 | 0.965182 | 11 | TCTTTCCTGCA | CTTATCTGTCCCCACCAG | - |
| GCR1#MA0304.1 | 0 | 0.0159771 | 7.60512 | 0.986704 | 8 | TCTTTCCTGCA | GGCTTCCA | - |
| SOK2#MA0385.1 | 0 | 0.0175243 | 8.34158 | 0.986704 | 11 | TCTTTCCTGCA | TGCCTGCAGGT | - |
| sd#MA0243.1 | 1 | 0.0176428 | 8.39798 | 0.986704 | 11 | TCTTTCCTGCA | GACATTCCTCGA | + |
| NFATC2#MA0152.1 | -1 | 0.0195062 | 9.28495 | 0.998691 | 7 | TCTTTCCTGCA | TTTTCCA | + |