MacroAPE 1083:CNhs13793 bl f2
From FANTOM5_SSTAR
Full Name: CNhs13793_bl_f2
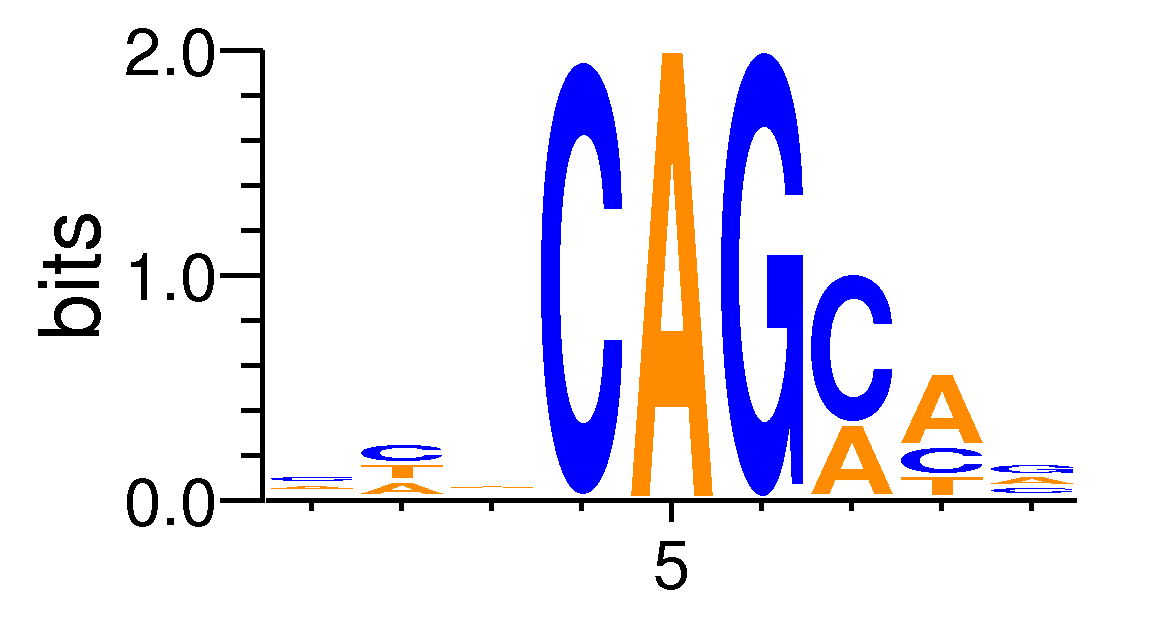
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| CUP2#MA0287.1 | -3 | 0.00144958 | 0.690002 | 0.756766 | 6 | CCTCAGCAG | CAGCAAAAATG | + |
| SWI5#MA0402.1 | 0 | 0.00229046 | 1.09026 | 0.756766 | 8 | CCTCAGCAG | AACCAGCA | - |
| NFE2L2#MA0150.1 | 3 | 0.00238477 | 1.13515 | 0.756766 | 8 | CCTCAGCAG | ATGACTCAGCA | + |
| ACE2#MA0267.1 | -1 | 0.00326037 | 1.55194 | 0.775969 | 7 | CCTCAGCAG | ACCAGCA | + |
| NHLH1#MA0048.1 | 0 | 0.0068987 | 3.28378 | 0.900321 | 9 | CCTCAGCAG | GCGCAGCTGCGT | + |
| Mafb#MA0117.1 | 1 | 0.00733585 | 3.49186 | 0.900321 | 7 | CCTCAGCAG | GCGTCAGC | - |
| BRCA1#MA0133.1 | -2 | 0.0086531 | 4.11888 | 0.900321 | 7 | CCTCAGCAG | ACAACAC | + |
| REST#MA0138.2 | -2 | 0.00882582 | 4.20109 | 0.900321 | 7 | CCTCAGCAG | TCAGCACCATGGACAGCGC | - |
| REST#MA0138.2 | -1 | 0.00945716 | 4.50161 | 0.900321 | 8 | CCTCAGCAG | TTCAGCACCATGGACAGCGCC | + |
| Tal1::Gata1#MA0140.1 | 9 | 0.0107809 | 5.13172 | 0.933041 | 9 | CCTCAGCAG | CTTATCTGTCCCCACCAG | - |
| CRZ1#MA0285.1 | -1 | 0.0141732 | 6.74645 | 1 | 8 | CCTCAGCAG | CTAAGCCAC | + |
| PHD1#MA0355.1 | 2 | 0.0146658 | 6.98094 | 1 | 8 | CCTCAGCAG | ACCTGCAGCA | + |
| Myf#MA0055.1 | 0 | 0.0180962 | 8.61377 | 1 | 9 | CCTCAGCAG | CAGCAGCTGCTG | + |
| SOK2#MA0385.1 | 2 | 0.0197242 | 9.3887 | 1 | 9 | CCTCAGCAG | ACCTGCAGGCA | + |