MacroAPE 1083:NR0B1 si
From FANTOM5_SSTAR
Full Name: NR0B1_si
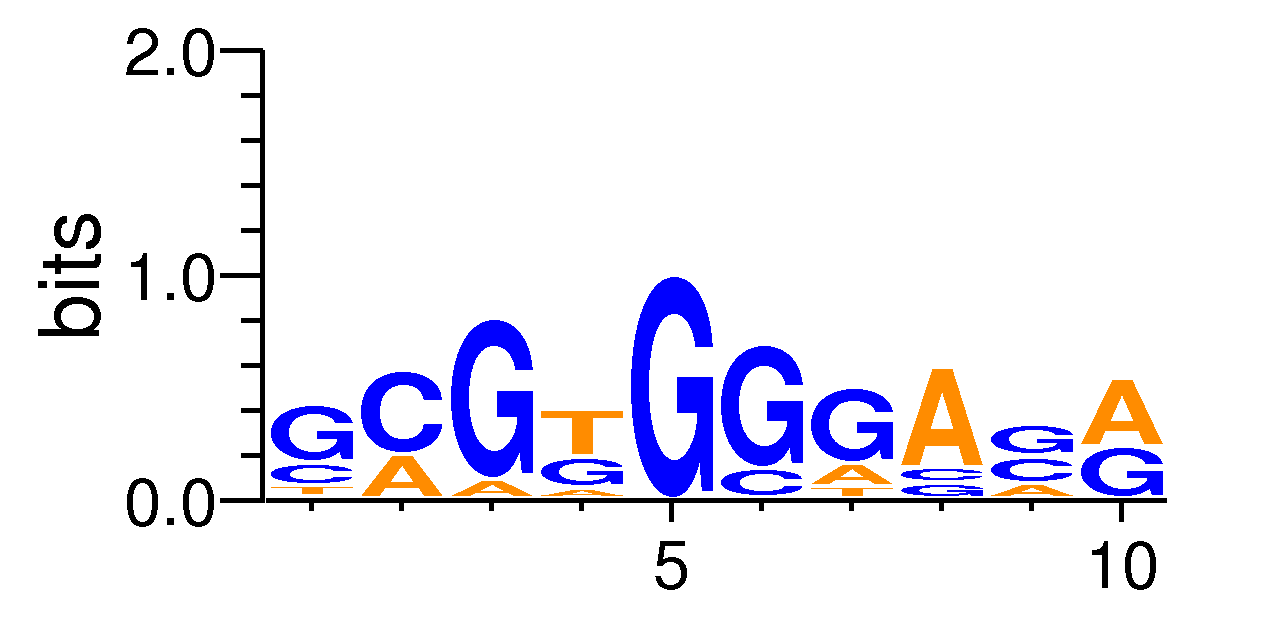
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| ZMS1#MA0441.1 | 1 | 0.000447223 | 0.212878 | 0.241293 | 8 | GCGTGGGAGA | TGCGGGGAA | - |
| Egr1#MA0162.1 | 1 | 0.000507582 | 0.241609 | 0.241293 | 10 | GCGTGGGAGA | TGCGTGGGCGT | + |
| MZF1_5-13#MA0057.1 | 1 | 0.00119179 | 0.567294 | 0.311918 | 9 | GCGTGGGAGA | GGAGGGGGAA | + |
| MIG1#MA0337.1 | 0 | 0.00152435 | 0.725591 | 0.311918 | 7 | GCGTGGGAGA | GCGGGGG | - |
| YPR022C#MA0436.1 | -1 | 0.00164037 | 0.780818 | 0.311918 | 7 | GCGTGGGAGA | CGTGGGG | - |
| NHP10#MA0344.1 | 0 | 0.00285735 | 1.3601 | 0.452773 | 8 | GCGTGGGAGA | GCCGGGGA | + |
| Su(H)#MA0085.1 | 0 | 0.0053629 | 2.55274 | 0.582982 | 10 | GCGTGGGAGA | CTGTGGGAAACGAGAT | + |
| SP1#MA0079.2 | -2 | 0.00544103 | 2.58993 | 0.582982 | 8 | GCGTGGGAGA | GGGGGCGGGG | - |
| Pax4#MA0068.1 | 1 | 0.00599225 | 2.85231 | 0.582982 | 10 | GCGTGGGAGA | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| MZF1_1-4#MA0056.1 | -2 | 0.00613179 | 2.91873 | 0.582982 | 6 | GCGTGGGAGA | TGGGGA | + |
| YML081W#MA0431.1 | 2 | 0.00714446 | 3.40076 | 0.617511 | 7 | GCGTGGGAGA | GTGCGGGGT | - |
| GAL4#MA0299.1 | -1 | 0.00969934 | 4.61689 | 0.67372 | 9 | GCGTGGGAGA | CGGGGAACAGTACTC | + |
| MIG2#MA0338.1 | 1 | 0.0106594 | 5.07389 | 0.67372 | 6 | GCGTGGGAGA | TGCGGGG | - |
| UGA3#MA0410.1 | 0 | 0.0113085 | 5.38284 | 0.67372 | 8 | GCGTGGGAGA | CGGCGGGA | + |
| MIG3#MA0339.1 | 1 | 0.0113223 | 5.3894 | 0.67372 | 6 | GCGTGGGAGA | TGCGGGG | - |
| Trl#MA0205.1 | 0 | 0.0113379 | 5.39682 | 0.67372 | 10 | GCGTGGGAGA | GAGAGAGCAA | - |
| SUT1#MA0399.1 | 1 | 0.0127576 | 6.07263 | 0.713492 | 6 | GCGTGGGAGA | CGCGGGG | + |
| RSC30#MA0375.1 | 1 | 0.0157502 | 7.49708 | 0.831919 | 7 | GCGTGGGAGA | CGCGCGCG | - |
| znf143#MA0088.1 | 8 | 0.0190794 | 9.08181 | 0.876505 | 10 | GCGTGGGAGA | GCAAGGCATGATGGGAAATC | - |
| PPARG::RXRA#MA0065.2 | 0 | 0.0196926 | 9.3737 | 0.876505 | 10 | GCGTGGGAGA | GTAGGGCAAAGGTCA | + |
| ADR1#MA0268.1 | 0 | 0.0202002 | 9.61531 | 0.876505 | 7 | GCGTGGGAGA | GTGGGGT | - |
| z#MA0255.1 | 2 | 0.0202819 | 9.6542 | 0.876505 | 8 | GCGTGGGAGA | TTGAGTGATT | + |