UBERON:0000926: Difference between revisions
From FANTOM5_SSTAR
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{UBERON | {{UBERON | ||
| | |alt_id=UBERON:0003263;;UBERON:FBbt_00000126-FMA_69072-MIAA_0000174-XAO_0000050-ZFA_0000041 | ||
|comment=Taxon notes: sponges do not seem to have a mesoderm and accordingly Amphimedon lacks transcription factors involved in mesoderm development (Fkh, Gsc, Twist, Snail)[http://www.nature.com/nature/journal/v466/n7307/full/nature09201.html]. Mesoderm may not be homologous across verteberates[UBERONREF:0000002] | |comment=Taxon notes: sponges do not seem to have a mesoderm and accordingly Amphimedon lacks transcription factors involved in mesoderm development (Fkh, Gsc, Twist, Snail)[http://www.nature.com/nature/journal/v466/n7307/full/nature09201.html]. Mesoderm may not be homologous across verteberates[UBERONREF:0000002] | ||
|created_by= | |created_by= | ||
| | |def="The middle germ layer of the embryo, between the endoderm and ectoderm." [Wikipedia:Mesoderm] | ||
|derives_from= | |derives_from= | ||
|develops_from=UBERON:0006603 | |develops_from=UBERON:0006603 | ||
|disjoint_from= | |||
|has_quality= | |has_quality= | ||
|id=UBERON:0000926 | |||
|is_a=UBERON:0000923 | |||
|is_obsolete= | |||
|located_in= | |located_in= | ||
|name=mesoderm | |||
|namespace= | |||
|obo_creation_date= | |||
|part_of= | |part_of= | ||
|preceded_by= | |||
|property_value=IAO:0000412 http://purl.obolibrary.org/obo/uberon.owl;;RO:0002161 NCBITaxon:6040;;UBPROP:0000001 "Primary germ layer that is the middle of the embryonic germ layers.[AAO]" xsd:string | |property_value=IAO:0000412 http://purl.obolibrary.org/obo/uberon.owl;;RO:0002161 NCBITaxon:6040;;UBPROP:0000001 "Primary germ layer that is the middle of the embryonic germ layers.[AAO]" xsd:string | ||
| | |subset=uberon_slim;;vertebrate_core | ||
| | |synonym="entire mesoderm" RELATED [SCTID:362854004, UBERON:cjm];;"mesodermal mantle" RELATED [] | ||
|union_of= | |||
|xref=AAO:0000304;;BILA:0000037;;BTO:0000839;;EFO:0001981;;EHDAA2:0001128;;EHDAA:124;;EHDAA:160;;EHDAA:168;;EHDAA:183;;EMAPA:16083;;EV:0100006;;FBbt:00000126;;FMA:69072;;GAID:522;;http://upload.wikimedia.org/wikipedia/commons/e/e8/Mesoderm.png;;http://upload.wikimedia.org/wikipedia/commons/thumb/e/e8/Mesoderm.png/200px-Mesoderm.png;;MAT:0000174;;MESH:A.16.254.425.660;;MIAA:0000174;;ncithesaurus:Mesoderm;;SCTID:362854004;;TAO:0000041;;VHOG:0000152;;XAO:0000050;;ZFA:0000041 | |||
}} | }} | ||
Revision as of 10:56, 12 September 2014
| Name: | mesoderm | ||
|---|---|---|---|
| Definition: | "The middle germ layer of the embryo, between the endoderm and ectoderm." [Wikipedia:Mesoderm] | ||
| Xrefs: |
| ||
| Synonyms: |
"entire mesoderm" RELATED [SCTID:362854004, UBERON:cjm] "mesodermal mantle" RELATED [] | ||
| Comments: | Taxon notes: sponges do not seem to have a mesoderm and accordingly Amphimedon lacks transcription factors involved in mesoderm development (Fkh, Gsc, Twist, Snail)[1]. Mesoderm may not be homologous across verteberates[UBERONREF:0000002] | ||
| Alt_id: |
UBERON:0003263 UBERON:FBbt_00000126-FMA_69072-MIAA_0000174-XAO_0000050-ZFA_0000041 | ||
| Subset: |
uberon_slim vertebrate_core |
Ontology association<br>Each term has an is_a parent in the Uberon Ontology, which has a linkage to an another entity and FANTOM5 samples.Libraries were grouped into mutually exclusive facets according to the FANTOM5 sample ontology mapping to UBERON ontologies.<br><br>link to ontology dataset<br>data
Parents
| is_a: | UBERON:0000923(germ layer) |
|---|---|
| develops_from: | UBERON:0006603(presumptive mesoderm) |
Children
| is a: | UBERON:0005721 (pronephric mesoderm) |
|---|---|
| derives from: | FF:0200026 (mouse mesoderm, embryonic day 8.5 sample) |
| develops from: | UBERON:0000042 (serous membrane),UBERON:0000077 (mixed endoderm/mesoderm-derived structure),UBERON:0000078 (mixed ectoderm/mesoderm/endoderm-derived structure),UBERON:0000990 (reproductive system),UBERON:0001136 (mesothelium),UBERON:0001235 (adrenal cortex),UBERON:0001986 (endothelium),UBERON:0002072 (hypodermis),UBERON:0002358 (peritoneum),UBERON:0002385 (muscle tissue),UBERON:0003064 (intermediate mesoderm),UBERON:0003068 (axial mesoderm),UBERON:0003081 (lateral plate mesoderm),UBERON:0004120 (mesoderm-derived structure),UBERON:0006904 (head mesenchyme from mesoderm),UBERON:0009617 (head paraxial mesoderm),UBERON:0009618 (trunk paraxial mesoderm),UBERON:0012275 (meso-epithelium) |
| part of: | CL:0000222 (mesodermal cell),UBERON:0003064 (intermediate mesoderm),UBERON:0003068 (axial mesoderm),UBERON:0003077 (paraxial mesoderm),UBERON:0003081 (lateral plate mesoderm),UBERON:0003082 (myotome),UBERON:0004016 (dermatome),UBERON:0004290 (dermomyotome),UBERON:0007285 (presumptive paraxial mesoderm) |
Ontology Tree: Loaded from BioPortal
Ontorolgy tree(Small window open)
FF samples<br>It includes FANTOM5 samples that overlay the Uberon ontology
Mouse (Mus musculus)
- 11947-126B3 (Mesoderm, embryo E8.5)
Enrichment analysis: top 100 FFCP enriched with this ontology term TOP 100 FANTOM5 Cage Peaks enriched with UBERON:0000926 (mesoderm), sorted by p-values <br>Analyst: Hideya Kawaji<br><br>link to source dataset <br>human : data <br>mouse : data
| P-value | FFCP | Short description |
|---|---|---|
| 6.38e-40 | FFCP PHASE2:Hg19::chrX:153637534..153637567,-#_ef840cec790a7a3714ebf8810e09dd8a | p1@DNASE1L1 |
| 6.38e-40 | FFCP PHASE1:Hg19::chrX:153637534..153637567,-#_ef840cec790a7a3714ebf8810e09dd8a | p1@DNASE1L1 |
| 6.15e-39 | FFCP PHASE2:Hg19::chr1:228270784..228270847,+#_6c13f00b64ddc6c31fe4632305b109f9 | p2@ARF1 |
| 6.15e-39 | FFCP PHASE1:Hg19::chr1:228270784..228270847,+#_6c13f00b64ddc6c31fe4632305b109f9 | p2@ARF1 |
| 9.61e-38 | FFCP PHASE2:Hg19::chr12:54812992..54813035,-#_220a11004d78d2fc2c33d36af42b9a61 | p1@ITGA5 |
| 9.61e-38 | FFCP PHASE1:Hg19::chr12:54812992..54813035,-#_220a11004d78d2fc2c33d36af42b9a61 | p1@ITGA5 |
| 1.56e-36 | FFCP PHASE2:Hg19::chr7:143078379..143078454,+#_bc25188a6e6b60c0373d19a0103a2306 | p1@ZYX |
| 1.56e-36 | FFCP PHASE1:Hg19::chr7:143078379..143078454,+#_bc25188a6e6b60c0373d19a0103a2306 | p1@ZYX |
| 2.82e-36 | FFCP PHASE2:Hg19::chr4:103747936..103747993,-#_102549c8bf1fa933fd4ba975dffeca4c | p6@UBE2D3 |
| 2.82e-36 | FFCP PHASE1:Hg19::chr4:103747936..103747993,-#_102549c8bf1fa933fd4ba975dffeca4c | p6@UBE2D3 |
| 5.32e-36 | FFCP PHASE1:Hg19::chr15:90358115..90358195,-#_f3b10efc769a32e8d79b1784ca462011 | p3@ANPEP |
| 5.32e-36 | FFCP PHASE2:Hg19::chr15:90358115..90358195,-#_f3b10efc769a32e8d79b1784ca462011 | p3@ANPEP |
| 8.45e-36 | FFCP PHASE1:Hg19::chr5:180229791..180229829,-#_d7f248739dcba6ec1f4cad2b2606733b | p1@MGAT1 |
| 8.45e-36 | FFCP PHASE2:Hg19::chr5:180229791..180229829,-#_d7f248739dcba6ec1f4cad2b2606733b | p1@MGAT1 |
| 1.6e-35 | FFCP PHASE2:Hg19::chr11:504446..504486,-#_fdbc7c00e226a25c64a28dad997cb2ca | p4@RNH1 |
| 1.6e-35 | FFCP PHASE1:Hg19::chr11:504446..504486,-#_fdbc7c00e226a25c64a28dad997cb2ca | p4@RNH1 |
| 2.85e-35 | FFCP PHASE2:Hg19::chr19:44174330..44174355,-#_9ef84cbd9b0d0b16223a42f380498b7e | p2@PLAUR |
| 2.85e-35 | FFCP PHASE1:Hg19::chr19:44174330..44174355,-#_9ef84cbd9b0d0b16223a42f380498b7e | p2@PLAUR |
| 3.37e-35 | FFCP PHASE1:Hg19::chr1:21616690..21616719,-#_902e9d06d5508c464ae4ec9549320d84 | p4@ECE1 |
| 3.37e-35 | FFCP PHASE2:Hg19::chr1:21616690..21616719,-#_902e9d06d5508c464ae4ec9549320d84 | p4@ECE1 |
| 5.03e-35 | FFCP PHASE1:Hg19::chr7:143079458..143079487,+#_2352b848aae5ed0261fe54b912196d14 | p4@ZYX |
| 5.03e-35 | FFCP PHASE2:Hg19::chr7:143079458..143079487,+#_2352b848aae5ed0261fe54b912196d14 | p4@ZYX |
| 1.17e-34 | FFCP PHASE1:Hg19::chr4:15704679..15704733,+#_705cd95c97795ba13103f06d938f84b3 | p1@BST1 |
| 1.17e-34 | FFCP PHASE2:Hg19::chr4:15704679..15704733,+#_705cd95c97795ba13103f06d938f84b3 | p1@BST1 |
| 1.75e-34 | FFCP PHASE2:Hg19::chrX:153637522..153637533,-#_5848209969245f1b446438bb574fd3e5 | p4@DNASE1L1 |
| 1.75e-34 | FFCP PHASE1:Hg19::chrX:153637522..153637533,-#_5848209969245f1b446438bb574fd3e5 | p4@DNASE1L1 |
| 1.89e-34 | FFCP PHASE1:Hg19::chr19:50015870..50015934,+#_3bfa8521e47c0bfacc3109b78f3114bb | p2@FCGRT |
| 1.89e-34 | FFCP PHASE2:Hg19::chr19:50015870..50015934,+#_3bfa8521e47c0bfacc3109b78f3114bb | p2@FCGRT |
| 2.1e-34 | FFCP PHASE1:Hg19::chr12:54813229..54813273,-#_e08a5b61753d71f83631ed75e2cbda8c | p3@ITGA5 |
| 2.1e-34 | FFCP PHASE2:Hg19::chr12:54813229..54813273,-#_e08a5b61753d71f83631ed75e2cbda8c | p3@ITGA5 |
| 4.33e-34 | FFCP PHASE1:Hg19::chr19:44174310..44174329,-#_fa5b0ab59b853e16f8cb8adbca538512 | p1@PLAUR |
| 4.33e-34 | FFCP PHASE2:Hg19::chr19:44174310..44174329,-#_fa5b0ab59b853e16f8cb8adbca538512 | p1@PLAUR |
| 7.52e-34 | FFCP PHASE2:Hg19::chr1:221052776..221052799,+#_a1446ba40352037a24d48cc7f7ef9d91 | p1@HLX |
| 7.52e-34 | FFCP PHASE1:Hg19::chr1:221052776..221052799,+#_a1446ba40352037a24d48cc7f7ef9d91 | p1@HLX |
| 8.49e-34 | FFCP PHASE2:Hg19::chr4:159093517..159093533,-#_8c85a728f89a11fd03bba24f3341e4cf | p1@FAM198B |
| 8.49e-34 | FFCP PHASE1:Hg19::chr4:159093517..159093533,-#_8c85a728f89a11fd03bba24f3341e4cf | p1@FAM198B |
| 3.78e-33 | FFCP PHASE1:Hg19::chr15:90358048..90358103,-#_0511f154ba60356ec188990130384b07 | p1@ANPEP |
| 3.78e-33 | FFCP PHASE2:Hg19::chr15:90358048..90358103,-#_0511f154ba60356ec188990130384b07 | p1@ANPEP |
| 3.32e-32 | FFCP PHASE2:Hg19::chr7:97911007..97911027,+#_26435938e3239e70d376d510cef8423a | p3@BRI3 |
| 3.32e-32 | FFCP PHASE1:Hg19::chr7:97911007..97911027,+#_26435938e3239e70d376d510cef8423a | p3@BRI3 |
| 5.72e-32 | FFCP PHASE1:Hg19::chr7:97910981..97911006,+#_100faa4865664d5161f520a0008f41e6 | p1@BRI3 |
| 5.72e-32 | FFCP PHASE2:Hg19::chr7:97910981..97911006,+#_100faa4865664d5161f520a0008f41e6 | p1@BRI3 |
| 5.73e-32 | FFCP PHASE2:Hg19::chr19:54371114..54371180,+#_d61cfb42500136991ccc245a081f2934 | p1@MYADM |
| 5.73e-32 | FFCP PHASE1:Hg19::chr19:54371114..54371180,+#_d61cfb42500136991ccc245a081f2934 | p1@MYADM |
| 7.25e-32 | FFCP PHASE2:Hg19::chr4:103747996..103748022,-#_90041e8a0ad2a882952d5083ea20fc85 | p4@UBE2D3 |
| 7.25e-32 | FFCP PHASE1:Hg19::chr4:103747996..103748022,-#_90041e8a0ad2a882952d5083ea20fc85 | p4@UBE2D3 |
| 7.82e-32 | FFCP PHASE1:Hg19::chr19:50015936..50015981,+#_cd91040758a5d2574d519cedd378f5e2 | p3@FCGRT |
| 7.82e-32 | FFCP PHASE2:Hg19::chr19:50015936..50015981,+#_cd91040758a5d2574d519cedd378f5e2 | p3@FCGRT |
| 8.33e-32 | FFCP PHASE2:Hg19::chr8:134584101..134584143,-#_fab013e5644acfadca752ea3bd0b898c | p2@ST3GAL1 |
| 8.33e-32 | FFCP PHASE1:Hg19::chr8:134584101..134584143,-#_fab013e5644acfadca752ea3bd0b898c | p2@ST3GAL1 |
| 1.49e-31 | FFCP PHASE1:Hg19::chr7:143078652..143078754,+#_80f55d84fa4d989503b80ebdc41c0e34 | p2@ZYX |
| 1.49e-31 | FFCP PHASE2:Hg19::chr7:143078652..143078754,+#_80f55d84fa4d989503b80ebdc41c0e34 | p2@ZYX |
| 2.42e-31 | FFCP PHASE1:Hg19::chr1:17944856..17944906,+#_d7151b05a4f1b1319895e485cabb2488 | p3@ARHGEF10L |
| 2.42e-31 | FFCP PHASE2:Hg19::chr1:17944856..17944906,+#_d7151b05a4f1b1319895e485cabb2488 | p3@ARHGEF10L |
| 2.88e-31 | FFCP PHASE1:Hg19::chr12:65153154..65153171,-#_6039359dceb2a7b1f40413f4ecdd75a2 | p2@GNS |
| 2.88e-31 | FFCP PHASE2:Hg19::chr12:65153154..65153171,-#_6039359dceb2a7b1f40413f4ecdd75a2 | p2@GNS |
| 8.04e-31 | FFCP PHASE2:Hg19::chr1:221053049..221053075,+#_ff7735fc58cd4e6d179c18ea5b02a783 | p3@HLX |
| 8.04e-31 | FFCP PHASE1:Hg19::chr1:221053049..221053075,+#_ff7735fc58cd4e6d179c18ea5b02a783 | p3@HLX |
| 8.77e-31 | FFCP PHASE2:Hg19::chr1:87170448..87170539,+#_6d84a4359abfd89df11fcd7bdb822c12 | p1@SH3GLB1 |
| 8.77e-31 | FFCP PHASE1:Hg19::chr1:87170448..87170539,+#_6d84a4359abfd89df11fcd7bdb822c12 | p1@SH3GLB1 |
| 9.41e-31 | FFCP PHASE1:Hg19::chr10:104403641..104403699,+#_f23d32b302a4971dcdd71b7d02eb07d5 | p@chr10:104403641..104403699, + |
| 9.41e-31 | FFCP PHASE2:Hg19::chr10:104403641..104403699,+#_f23d32b302a4971dcdd71b7d02eb07d5 | p@chr10:104403641..104403699, + |
| 9.71e-31 | FFCP PHASE2:Hg19::chr4:80994210..80994284,-#_c2af42866f36622f2a099a2888441e60 | p2@ANTXR2 |
| 9.71e-31 | FFCP PHASE1:Hg19::chr4:80994210..80994284,-#_c2af42866f36622f2a099a2888441e60 | p2@ANTXR2 |
| 9.83e-31 | FFCP PHASE2:Hg19::chr4:122618095..122618161,-#_016568157f0bdad1fa07b37c8b31fb49 | p1@ANXA5 |
| 9.83e-31 | FFCP PHASE1:Hg19::chr4:122618095..122618161,-#_016568157f0bdad1fa07b37c8b31fb49 | p1@ANXA5 |
| 1.49e-30 | FFCP PHASE1:Hg19::chr9:130616952..130616973,-#_b7a543cd39b1db02aaf4b5fd1c35f5b5 | p3@ENG |
| 1.49e-30 | FFCP PHASE2:Hg19::chr9:130616952..130616973,-#_b7a543cd39b1db02aaf4b5fd1c35f5b5 | p3@ENG |
| 1.87e-30 | FFCP PHASE1:Hg19::chr1:20987936..20987969,-#_e35b513ee7cd2296a23b3aa521c3fb62 | p2@DDOST |
| 1.87e-30 | FFCP PHASE2:Hg19::chr1:20987936..20987969,-#_e35b513ee7cd2296a23b3aa521c3fb62 | p2@DDOST |
| 3.28e-30 | FFCP PHASE2:Hg19::chrX:46433193..46433237,+#_3864dfe743d7e7088fe5ea47c54c2b66 | p1@CHST7 |
| 3.28e-30 | FFCP PHASE1:Hg19::chrX:46433193..46433237,+#_3864dfe743d7e7088fe5ea47c54c2b66 | p1@CHST7 |
| 3.51e-30 | FFCP PHASE1:Hg19::chr17:81037473..81037498,+#_ec3e97853942bad3cced97866597fb57 | p1@METRNL |
| 3.51e-30 | FFCP PHASE2:Hg19::chr17:81037473..81037498,+#_ec3e97853942bad3cced97866597fb57 | p1@METRNL |
| 3.8e-30 | FFCP PHASE1:Hg19::chr6:146864960..146864999,+#_b453996d2df6bbb53234ed380bd561bb | p1@RAB32 |
| 3.8e-30 | FFCP PHASE2:Hg19::chr6:146864960..146864999,+#_b453996d2df6bbb53234ed380bd561bb | p1@RAB32 |
| 5.98e-30 | FFCP PHASE1:Hg19::chr3:50273625..50273655,+#_8e06489cf03982f4df517860e79d8c00 | p1@GNAI2 |
| 5.98e-30 | FFCP PHASE2:Hg19::chr3:50273625..50273655,+#_8e06489cf03982f4df517860e79d8c00 | p1@GNAI2 |
| 6.52e-30 | FFCP PHASE1:Hg19::chr7:143077956..143078096,-#_963879b71e8c7e9cb80b02b9999713e3 | p1@ENST00000429630 |
| 6.52e-30 | FFCP PHASE2:Hg19::chr7:143077956..143078096,-#_963879b71e8c7e9cb80b02b9999713e3 | p1@ENST00000429630 |
| 7.62e-30 | FFCP PHASE2:Hg19::chr9:35732119..35732199,-#_972538a1a3f36d9b23cae1dfce250b06 | p1@TLN1 |
| 7.62e-30 | FFCP PHASE1:Hg19::chr9:35732119..35732199,-#_972538a1a3f36d9b23cae1dfce250b06 | p1@TLN1 |
| 9.35e-30 | FFCP PHASE1:Hg19::chr5:114938090..114938226,-#_4aabd60a6a325b6f9d59e5e8f1120629 | p1@TICAM2, p2@TMED7-TICAM2 |
| 9.35e-30 | FFCP PHASE2:Hg19::chr5:114938090..114938226,-#_4aabd60a6a325b6f9d59e5e8f1120629 | p1@TICAM2, p2@TMED7-TICAM2 |
| 1.51e-29 | FFCP PHASE1:Hg19::chr1:59762730..59762773,+#_cd8c85a2db32daafd75499928599070f | p1@FGGY |
| 1.51e-29 | FFCP PHASE2:Hg19::chr1:59762730..59762773,+#_cd8c85a2db32daafd75499928599070f | p1@FGGY |
| 1.57e-29 | FFCP PHASE2:Hg19::chr11:67120956..67120972,-#_6b4ce1c12cd94cdbe93f18c7a3ef0f4d | p2@POLD4 |
| 1.57e-29 | FFCP PHASE1:Hg19::chr11:67120956..67120972,-#_6b4ce1c12cd94cdbe93f18c7a3ef0f4d | p2@POLD4 |
| 1.96e-29 | FFCP PHASE1:Hg19::chr11:504867..504925,-#_a7db9716dd0988b362c9702063a72b24 | p3@RNH1 |
| 1.96e-29 | FFCP PHASE2:Hg19::chr11:504867..504925,-#_a7db9716dd0988b362c9702063a72b24 | p3@RNH1 |
| 3.49e-29 | FFCP PHASE1:Hg19::chr4:159092525..159092574,-#_2ce8cf32429c5fc912b829bfe47ec407 | p4@FAM198B |
| 3.49e-29 | FFCP PHASE2:Hg19::chr4:159092525..159092574,-#_2ce8cf32429c5fc912b829bfe47ec407 | p4@FAM198B |
| 5.21e-29 | FFCP PHASE2:Hg19::chr3:148709310..148709337,+#_f08f810aec085ad6fe464433a8630549 | p1@GYG1 |
| 5.21e-29 | FFCP PHASE1:Hg19::chr3:148709310..148709337,+#_f08f810aec085ad6fe464433a8630549 | p1@GYG1 |
| 6.23e-29 | FFCP PHASE1:Hg19::chr8:146078927..146078951,-#_91cf648a38ab4db34710588de59827cc | p4@COMMD5 |
| 6.23e-29 | FFCP PHASE2:Hg19::chr8:146078927..146078951,-#_91cf648a38ab4db34710588de59827cc | p4@COMMD5 |
| 7.59e-29 | FFCP PHASE2:Hg19::chr4:10118348..10118395,-#_37e2a6f50ca407a63a9601f89740b2a5 | p1@WDR1 |
| 7.59e-29 | FFCP PHASE1:Hg19::chr4:10118348..10118395,-#_37e2a6f50ca407a63a9601f89740b2a5 | p1@WDR1 |
| 8.57e-29 | FFCP PHASE1:Hg19::chr12:53694132..53694153,-#_a18b8dec27dce98f5093bf2092377fd8 | p1@ENST00000550263 |
| 8.57e-29 | FFCP PHASE2:Hg19::chr12:53694132..53694153,-#_a18b8dec27dce98f5093bf2092377fd8 | p1@ENST00000550263 |
Property "Property value" (as page type) with input value "UBPROP:0000001 "Primary germ layer that is the middle of the embryonic germ layers.[AAO]" xsd:string" contains invalid characters or is incomplete and therefore can cause unexpected results during a query or annotation process.
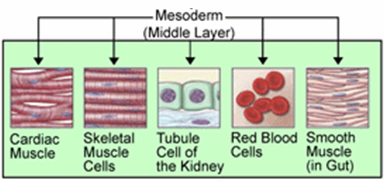 Mesoderm.png]
Mesoderm.png] 200px-Mesoderm.png]
200px-Mesoderm.png]