Introduction to FANTOM6
The FANTOM (Functional ANnoTation Of the Mammalian genome) consortium is a global collaborative effort dedicated to identifying and characterizing all functional elements within mammalian genomes. Our and others previous work has revealed a vast repertoire of long non-coding RNAs (lncRNAs) that are pervasively transcribed but lack protein-coding potential. FANTOM6 aims to systematically elucidate the origins and diverse regulatory roles of non-coding elements—including promoters, enhancers, and lncRNAs—within the human genome.
FANTOM6 Phase 1: High-throughput Functional Screening of lncRNAs
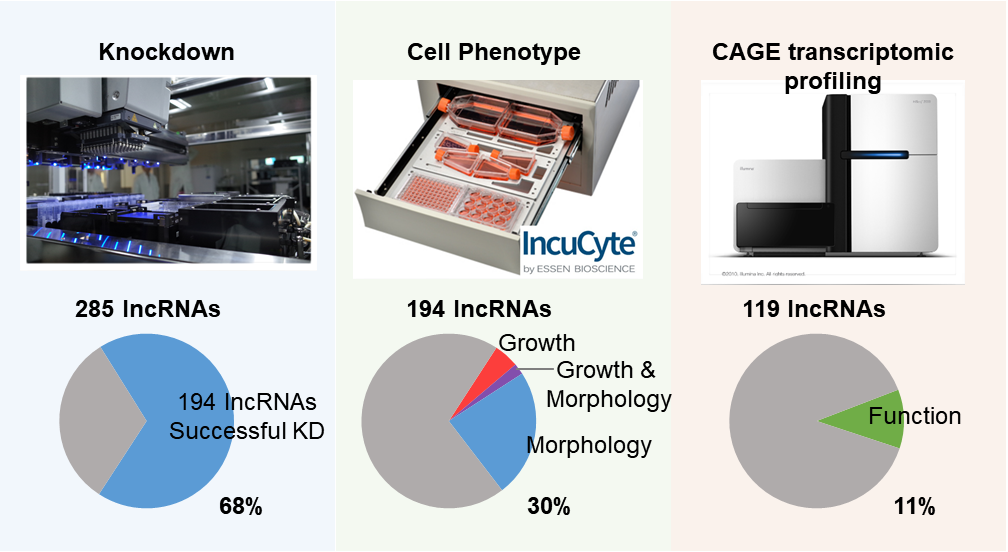
Building on the extensive catalog of lncRNAs identified in FANTOM5, the transcriptional landscape of the human genome was outlined. Hundreds of lncRNAs were selected for functional perturbation in two efficiently proliferating cell types—human dermal fibroblasts and induced pluripotent stem cells. Perturbations were performed at the RNA level using GapmeR antisense oligonucleotides, thereby specifically targeting RNA molecules without causing genomic alterations. Cellular and molecular phenotypes were systematically characterized through real-time imaging and CAGE-based transcriptomic profiling, revealing insights into the diverse regulatory functions of lncRNAs1-4.
FANTOM6 Phase 2: Interactome
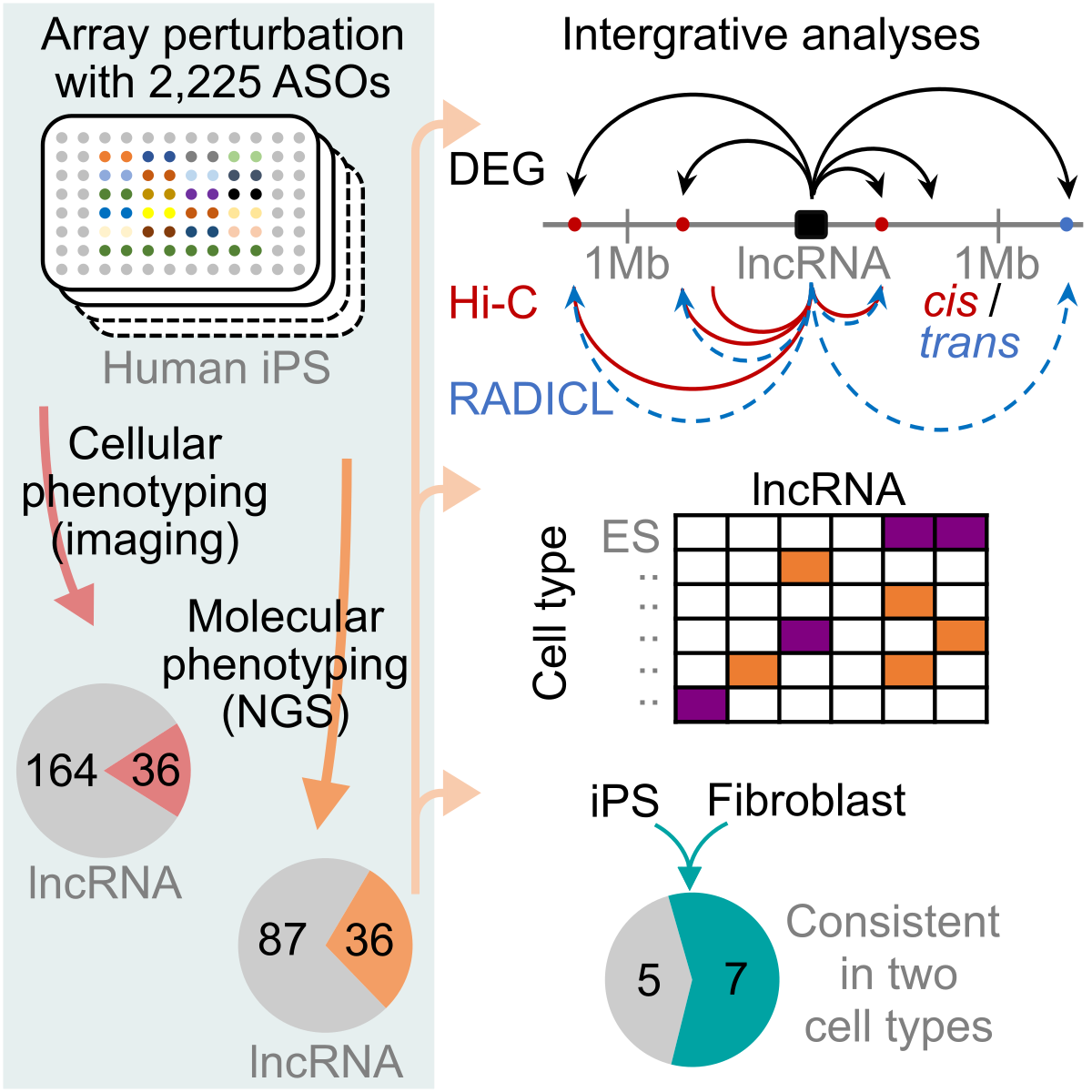
The second phase of FANTOM6 focuses on elucidating the functions of non-coding elements by examining their physical interactions with the genome and transcriptome. To obtain a comprehensive catalog of lncRNAs and eRNAs, we employed CAGE and cap-trapped full-length cDNA long-read sequencing (CFC-seq) to identify cis-regulatory elements and novel lncRNAs during the differentiation of induced pluripotent stem (iPS) cells into cortical neurons, and of monocytes into macrophages, as well as across a broader panel of human cells. Physical interactions among RNA–DNA, RNA–RNA, and DNA–DNA were mapped using RADICL-seq, PARIS-seq, and Hi-C, respectively. To provide functional context for these interactions, technologies such as scRNA-seq, scATAC-seq, CUT&Tag, and CRISPRi-based genetic screening were applied to selected cellular systems.

References
- Ramilowski, J. A. et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res 30, 1060–1072 (2020).
- Yip, C. W. et al. Antisense-oligonucleotide-mediated perturbation of long non-coding RNA reveals functional features in stem cells and across cell types. Cell Rep. 41, 111893 (2022).
- Agrawal, S. et al. Annotation of nuclear lncRNAs based on chromatin interactions. PLOS ONE 19, e0295971 (2024).
- Nobusada, T. et al. Update of the FANTOM web resource: enhancement for studying noncoding genomes. Nucleic Acids Res. 53, D419–D424 (2025).
Views and data
FANTOM6 Analysis Viewer with ZENBU-Reports
The view provides an interface to interactively access to analysis results performed in the FANTOM6 project. It is implemented with ZENBU-Report.
FANTOM6 Experiment Index
The view provides an interface to search for experiments and produced raw data files by the experiments.
FANTOM6 Data download site
The data files including sequences, mapping results, expression tables obtained in the FANTOM6 project are available via the FANTOM6 web site.
How to cite
In addition to relevant papers, please cite the following paper for using data obtained from this site:Nobusada T, et al. Update of the FANTOM web resource: enhancement for studying noncoding genomes. Nucleic Acids Res 53: D419–D424 (2025). 10.1093/nar/gkae1047