MacroAPE 1083:10-VGGAAACGAC
From FANTOM5_SSTAR
Full Name: 10-VGGAAACGAC,CNhs13491,BestGuess:REL_do_HM09(0.735873087466282),T:323.0(10.71%),B:2330.8(5.04%),P:1e-35
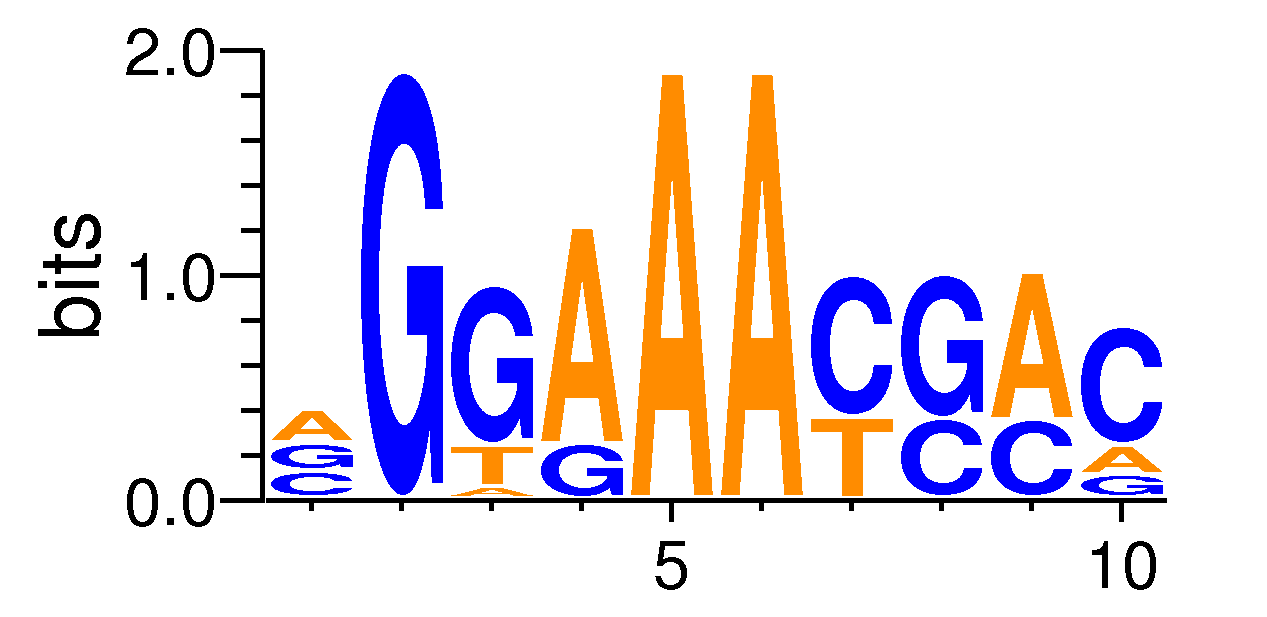
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| REL#MA0101.1 | -1 | 0.00065666 | 0.31257 | 0.351426 | 9 | AGGAAACGAC | GGAAATCCCC | - |
| RELA#MA0107.1 | -1 | 0.00079713 | 0.379434 | 0.351426 | 9 | AGGAAACGAC | GGAAATTCCC | - |
| Su(H)#MA0085.1 | 4 | 0.0017107 | 0.814295 | 0.351426 | 10 | AGGAAACGAC | CTGTGGGAAACGAGAT | + |
| STAT1#MA0137.2 | 0 | 0.0021196 | 1.00893 | 0.351426 | 10 | AGGAAACGAC | GGAAAACGAAACTG | + |
| dl_2#MA0023.1 | -1 | 0.00213075 | 1.01424 | 0.351426 | 9 | AGGAAACGAC | GGAAAACCCC | - |
| GABPA#MA0062.2 | 1 | 0.00239401 | 1.13955 | 0.351426 | 10 | AGGAAACGAC | CCGGAAGTGGC | + |
| dl_1#MA0022.1 | 0 | 0.00293421 | 1.39668 | 0.351426 | 10 | AGGAAACGAC | CGGAAAAACCCC | - |
| ETS1#MA0098.1 | 0 | 0.00297678 | 1.41695 | 0.351426 | 6 | AGGAAACGAC | CGGAAA | - |
| NFKB1#MA0105.1 | 0 | 0.00337912 | 1.60846 | 0.351979 | 10 | AGGAAACGAC | GGGGAATCCCC | - |
| STAT1#MA0137.2 | 7 | 0.00372683 | 1.77397 | 0.351979 | 8 | AGGAAACGAC | GGTTTCCGGGAAATG | - |
| FEV#MA0156.1 | 1 | 0.00494166 | 2.35223 | 0.424284 | 7 | AGGAAACGAC | CAGGAAAT | + |
| NF-kappaB#MA0061.1 | -1 | 0.00558631 | 2.65908 | 0.428318 | 9 | AGGAAACGAC | GGAAATTCCC | - |
| YRM1#MA0438.1 | 1 | 0.00595769 | 2.83586 | 0.428318 | 7 | AGGAAACGAC | ACGGAAAT | + |
| En1#MA0027.1 | -1 | 0.00689685 | 3.2829 | 0.428318 | 9 | AGGAAACGAC | GAACACTACTT | - |
| che-1#MA0260.1 | -2 | 0.00741051 | 3.5274 | 0.428318 | 6 | AGGAAACGAC | GAAACC | + |
| SPT23#MA0388.1 | -2 | 0.00757933 | 3.60776 | 0.428318 | 8 | AGGAAACGAC | GAAATCAA | + |
| SPI1#MA0080.2 | 0 | 0.0101808 | 4.84607 | 0.534179 | 6 | AGGAAACGAC | CGGAAG | + |
| Eip74EF#MA0026.1 | 1 | 0.0133622 | 6.3604 | 0.622971 | 6 | AGGAAACGAC | CCGGAAG | + |
| ELF5#MA0136.1 | 1 | 0.0139338 | 6.63247 | 0.622971 | 8 | AGGAAACGAC | AAGGAAGTA | - |
| CEP3#MA0282.1 | 2 | 0.0148458 | 7.0666 | 0.622971 | 6 | AGGAAACGAC | CTCGGAAA | + |
| GCR2#MA0305.1 | 0 | 0.0149651 | 7.1234 | 0.622971 | 7 | AGGAAACGAC | AGGAAGC | - |
| id1#MA0120.1 | 0 | 0.0153981 | 7.3295 | 0.622971 | 10 | AGGAAACGAC | CGAAAAGGAAAA | - |
| SPIB#MA0081.1 | 2 | 0.0158308 | 7.53546 | 0.622971 | 5 | AGGAAACGAC | AGAGGAA | + |
| UPC2#MA0411.1 | -2 | 0.0197729 | 9.4119 | 0.695575 | 7 | AGGAAACGAC | TATACGA | + |
| SPI1#MA0080.2 | 0 | 0.0208944 | 9.94572 | 0.695575 | 7 | AGGAAACGAC | AGGAAGT | + |
| STE12#MA0393.1 | -1 | 0.0208944 | 9.94572 | 0.695575 | 7 | AGGAAACGAC | TGAAACA | + |