MacroAPE 1083:AAACCCCCCC,motif448
From FANTOM5_SSTAR
Full Name: AAACCCCCCC,motif448,CNhs10859,T:1212.0(25.67%),B:14114.0(18.58%),P:1e-32
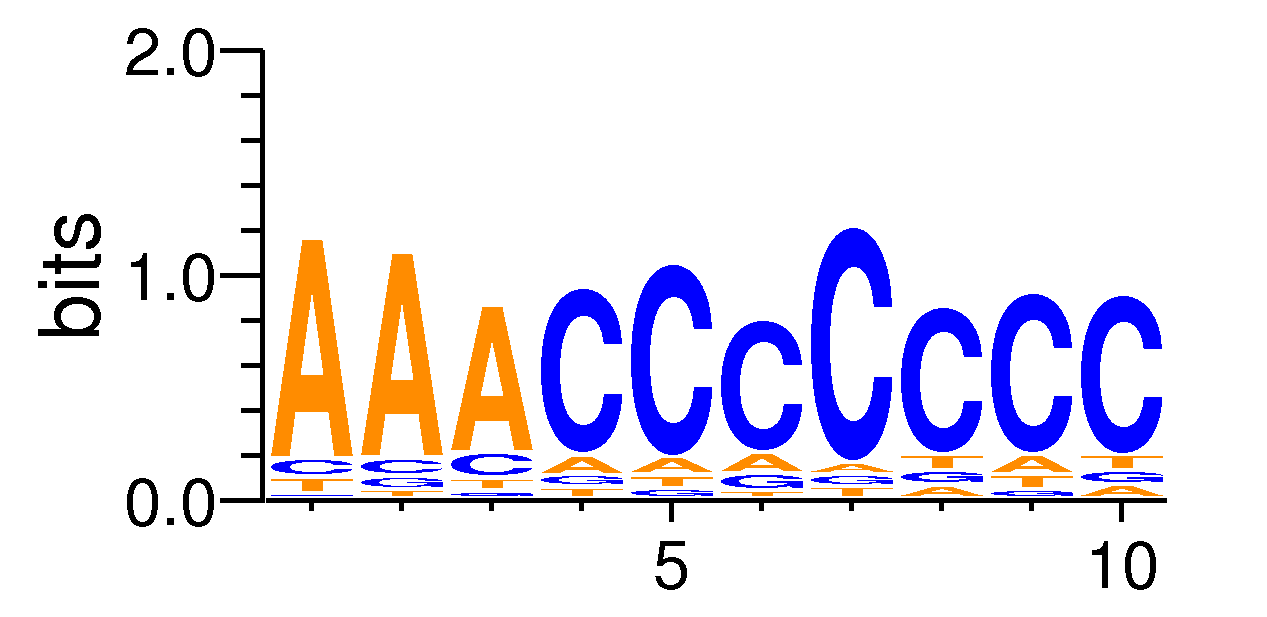
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| RREB1#MA0073.1 | 4 | 0.000396444 | 0.188708 | 0.358219 | 10 | AAACCCCCCC | CCCCAAACCACCCCCCCCCC | + |
| SP1#MA0079.2 | 0 | 0.00175541 | 0.835574 | 0.358219 | 10 | AAACCCCCCC | ACCCCGCCCC | - |
| Macho-1#MA0118.1 | -1 | 0.00181268 | 0.862836 | 0.358219 | 9 | AAACCCCCCC | GAACCCCCA | - |
| REL#MA0101.1 | 2 | 0.00284215 | 1.35286 | 0.358219 | 8 | AAACCCCCCC | GGAAATCCCC | - |
| MIG2#MA0338.1 | -3 | 0.00310438 | 1.47768 | 0.358219 | 7 | AAACCCCCCC | CCCCGCA | + |
| Pax4#MA0068.1 | 19 | 0.00316136 | 1.50481 | 0.358219 | 10 | AAACCCCCCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| MIG1#MA0337.1 | -3 | 0.00326571 | 1.55448 | 0.358219 | 7 | AAACCCCCCC | CCCCCGC | + |
| DAL81#MA0290.1 | 0 | 0.00359135 | 1.70948 | 0.358219 | 10 | AAACCCCCCC | AATCCCGCCCGCGGCTTTT | - |
| ADR1#MA0268.1 | -2 | 0.00360702 | 1.71694 | 0.358219 | 7 | AAACCCCCCC | ACCCCAC | + |
| dl_2#MA0023.1 | 2 | 0.00408577 | 1.94482 | 0.358219 | 8 | AAACCCCCCC | GGAAAACCCC | - |
| dl_1#MA0022.1 | 3 | 0.00416653 | 1.98327 | 0.358219 | 9 | AAACCCCCCC | CGGAAAAACCCC | - |
| RELA#MA0107.1 | 2 | 0.0054367 | 2.58787 | 0.415151 | 8 | AAACCCCCCC | GGAAATTCCC | - |
| MSN2#MA0341.1 | -3 | 0.00570667 | 2.71637 | 0.415151 | 5 | AAACCCCCCC | CCCCT | - |
| MIG3#MA0339.1 | -3 | 0.0070377 | 3.34995 | 0.443717 | 7 | AAACCCCCCC | CCCCGCA | + |
| YPR022C#MA0436.1 | -3 | 0.0070377 | 3.34995 | 0.443717 | 7 | AAACCCCCCC | CCCCACG | + |
| SP1#MA0079.2 | -3 | 0.0100941 | 4.80477 | 0.59664 | 7 | AAACCCCCCC | CCCCGCCCCC | + |
| CHA4#MA0283.1 | -2 | 0.0134619 | 6.40785 | 0.691347 | 8 | AAACCCCCCC | TCTCCGCC | - |
| btd#MA0443.1 | -1 | 0.0139843 | 6.65652 | 0.691347 | 9 | AAACCCCCCC | TCCGCCCCCT | - |
| NFKB1#MA0105.1 | 3 | 0.0141706 | 6.74522 | 0.691347 | 8 | AAACCCCCCC | GGGGAATCCCC | - |
| opa#MA0456.1 | -1 | 0.0146204 | 6.95932 | 0.691347 | 9 | AAACCCCCCC | GACCCCCCGCTG | + |
| YML081W#MA0431.1 | -2 | 0.0164874 | 7.84801 | 0.712703 | 8 | AAACCCCCCC | ACCCCGCAC | + |
| AFT2#MA0270.1 | -1 | 0.0167989 | 7.99626 | 0.712703 | 8 | AAACCCCCCC | CACACCCC | + |
| USV1#MA0413.1 | 0 | 0.0173328 | 8.25043 | 0.712703 | 10 | AAACCCCCCC | AAATTCCCCCTGAATTTGTG | + |
| MSN4#MA0342.1 | -3 | 0.0182116 | 8.6687 | 0.717633 | 5 | AAACCCCCCC | CCCCT | - |