MacroAPE 1083:CNhs13060 bl f1
From FANTOM5_SSTAR
Full Name: CNhs13060_bl_f1
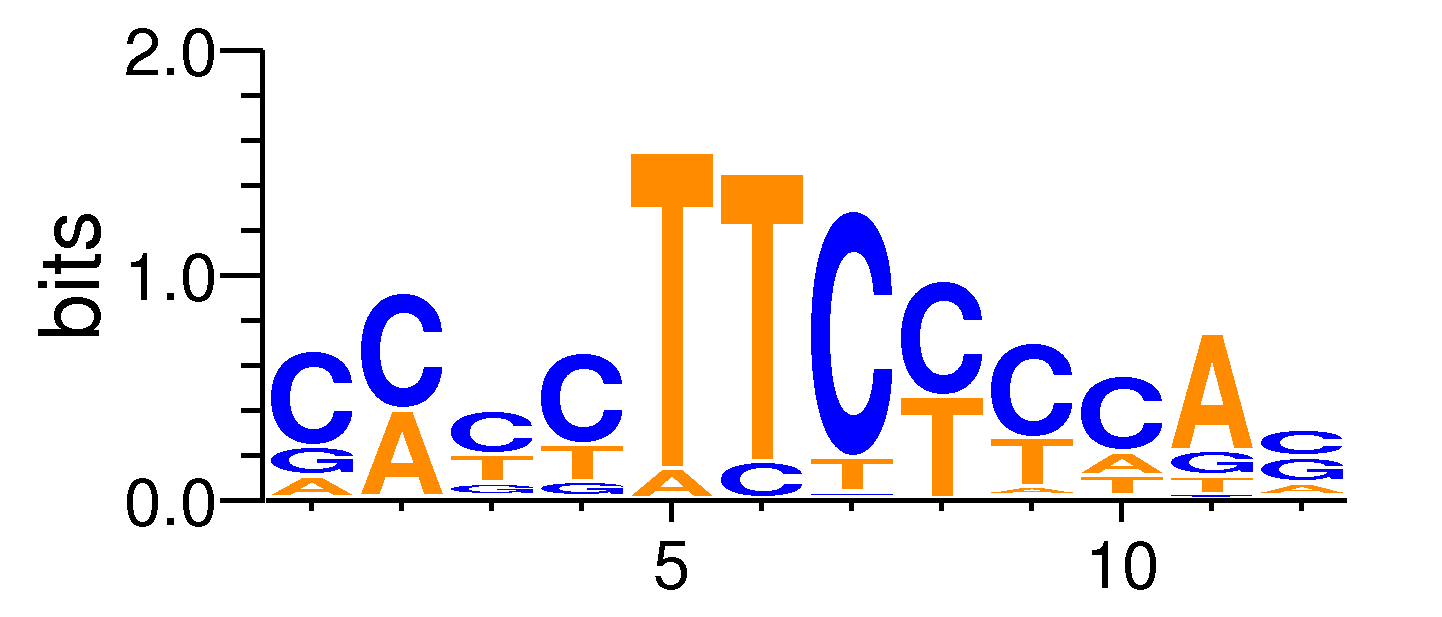
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| PPARG::RXRA#MA0065.2 | 3 | 0.00027902 | 0.132814 | 0.137522 | 12 | CCCCTTCCCCAC | TGACCTTTGCCCTAC | - |
| Klf4#MA0039.1 | -1 | 0.000434396 | 0.206772 | 0.137522 | 10 | CCCCTTCCCCAC | CCTTCCTTTA | - |
| SP1#MA0079.2 | 0 | 0.000434396 | 0.206772 | 0.137522 | 10 | CCCCTTCCCCAC | CCCCGCCCCC | + |
| SPI1#MA0080.1 | -3 | 0.00127699 | 0.607845 | 0.303204 | 6 | CCCCTTCCCCAC | CTTCCG | - |
| MZF1_1-4#MA0056.1 | -5 | 0.00265107 | 1.26191 | 0.503569 | 6 | CCCCTTCCCCAC | TCCCCA | - |
| Pax4#MA0068.1 | 9 | 0.00366462 | 1.74436 | 0.580079 | 12 | CCCCTTCCCCAC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| SPIB#MA0081.1 | -4 | 0.00437411 | 2.08207 | 0.593472 | 7 | CCCCTTCCCCAC | TTCCTCT | - |
| GCR2#MA0305.1 | -2 | 0.00900599 | 4.28685 | 0.997635 | 7 | CCCCTTCCCCAC | GCTTCCT | + |
| ZMS1#MA0441.1 | -4 | 0.0133067 | 6.334 | 0.997635 | 8 | CCCCTTCCCCAC | TTCCCCGCA | + |
| NFATC2#MA0152.1 | -4 | 0.0138535 | 6.59425 | 0.997635 | 7 | CCCCTTCCCCAC | TTTTCCA | + |
| RDR1#MA0360.1 | -3 | 0.0146952 | 6.9949 | 0.997635 | 8 | CCCCTTCCCCAC | GTTCCGCA | - |
| Ar#MA0007.1 | 10 | 0.0167921 | 7.99303 | 0.997635 | 12 | CCCCTTCCCCAC | ATAAGAACACCCTGTACCCGCC | + |
| TEAD1#MA0090.1 | 0 | 0.0201047 | 9.56985 | 0.997635 | 12 | CCCCTTCCCCAC | CACATTCCTCCG | + |
| ETS1#MA0098.1 | -3 | 0.0205557 | 9.78451 | 0.997635 | 6 | CCCCTTCCCCAC | CTTCCG | + |