MacroAPE 1083:EGR1 K562-EGR1-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: EGR1_K562-EGR1-ChIP-Seq/Homer
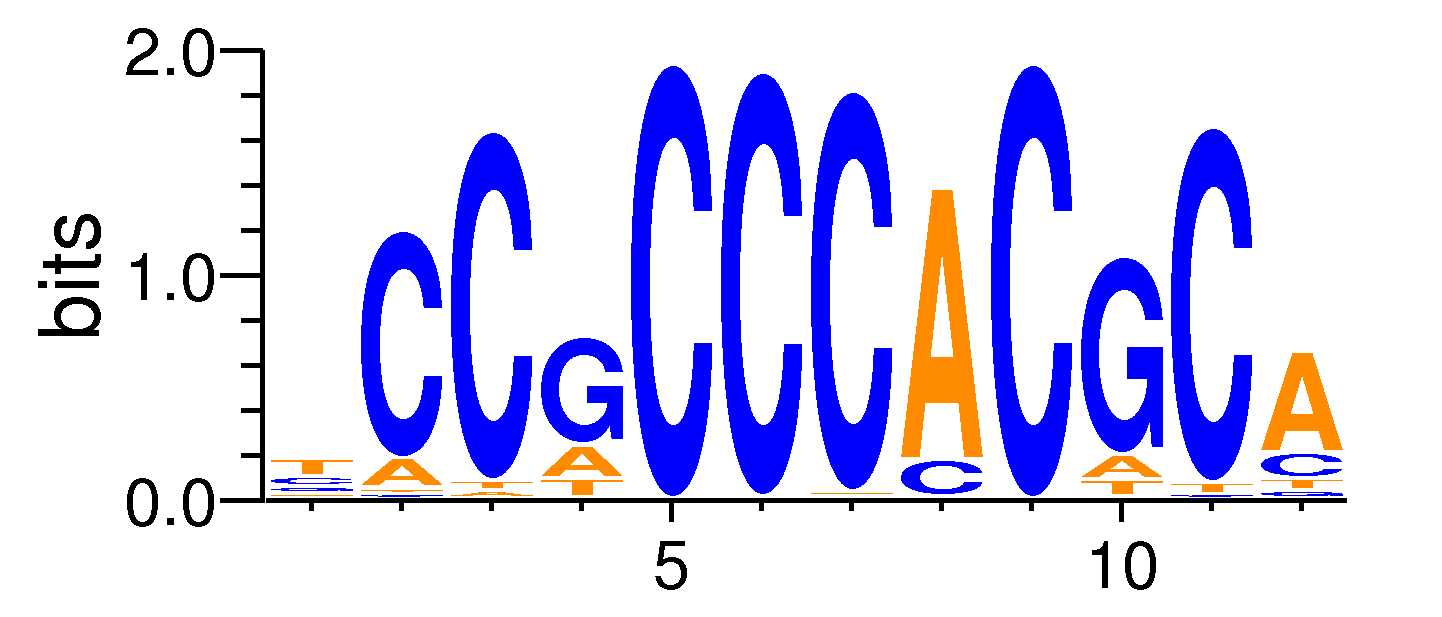
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Egr1#MA0162.1 | -1 | 1.50623e-08 | 7.16968e-06 | 1.43206e-05 | 11 | TCCGCCCACGCA | CCGCCCACGCA | - |
| btd#MA0443.1 | 0 | 0.000202828 | 0.0965461 | 0.0964197 | 10 | TCCGCCCACGCA | TCCGCCCCCT | - |
| Arnt::Ahr#MA0006.1 | -6 | 0.000638403 | 0.30388 | 0.202321 | 6 | TCCGCCCACGCA | CACGCA | - |
| RAP1#MA0359.1 | -2 | 0.00221677 | 1.05518 | 0.475467 | 10 | TCCGCCCACGCA | CACCCATACA | + |
| SP1#MA0079.2 | 1 | 0.00250048 | 1.19023 | 0.475467 | 9 | TCCGCCCACGCA | CCCCGCCCCC | + |
| YPR022C#MA0436.1 | -3 | 0.0043126 | 2.0528 | 0.650187 | 7 | TCCGCCCACGCA | CCCCACG | + |
| MIG3#MA0339.1 | -5 | 0.00522149 | 2.48543 | 0.650187 | 7 | TCCGCCCACGCA | CCCCGCA | + |
| MIG2#MA0338.1 | -5 | 0.00547092 | 2.60416 | 0.650187 | 7 | TCCGCCCACGCA | CCCCGCA | + |
| MIG1#MA0337.1 | -4 | 0.00751652 | 3.57787 | 0.79404 | 7 | TCCGCCCACGCA | CCCCCGC | + |
| DAL81#MA0290.1 | 3 | 0.00917638 | 4.36796 | 0.872448 | 12 | TCCGCCCACGCA | AATCCCGCCCGCGGCTTTT | - |
| Zfx#MA0146.1 | 0 | 0.010439 | 4.96894 | 0.902262 | 12 | TCCGCCCACGCA | GGGGCCGAGGCCTG | + |
| RREB1#MA0073.1 | 6 | 0.0116691 | 5.55448 | 0.924535 | 12 | TCCGCCCACGCA | CCCCAAACCACCCCCCCCCC | + |
| Klf4#MA0039.2 | 2 | 0.0139833 | 6.65607 | 0.957301 | 8 | TCCGCCCACGCA | GCCCCACCCA | - |
| ZMS1#MA0441.1 | -3 | 0.0140964 | 6.70989 | 0.957301 | 9 | TCCGCCCACGCA | TTCCCCGCA | + |
| YGR067C#MA0425.1 | 2 | 0.0178359 | 8.4899 | 0.998691 | 12 | TCCGCCCACGCA | ACCCCACTTTTTTG | + |
| YML081W#MA0431.1 | -4 | 0.0187979 | 8.94781 | 0.998691 | 8 | TCCGCCCACGCA | ACCCCGCAC | + |
| IME1#MA0320.1 | -1 | 0.0194652 | 9.26545 | 0.998691 | 8 | TCCGCCCACGCA | CCGCCGAG | + |
| INSM1#MA0155.1 | 0 | 0.0208057 | 9.90352 | 0.998691 | 12 | TCCGCCCACGCA | CGCCCCCTGACA | - |