MacroAPE 1083:GCAGGAAACCACAG,motif220
From FANTOM5_SSTAR
Full Name: GCAGGAAACCACAG,motif220,CNhs12575,T:1323.0(26.20%),B:13935.0(18.59%),P:1e-39
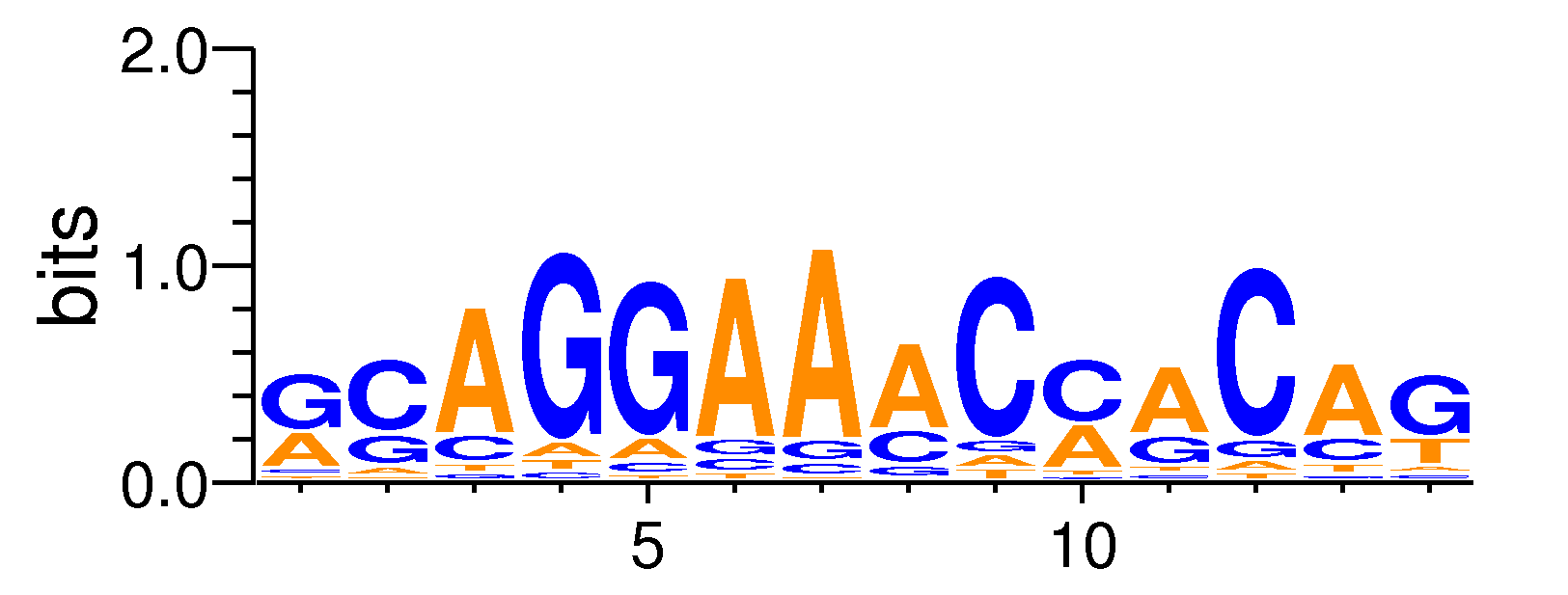
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| FEV#MA0156.1 | -1 | 0.00158097 | 0.752541 | 0.728824 | 8 | GCAGGAAACCACAG | CAGGAAAT | + |
| RUNX1#MA0002.2 | -5 | 0.00161585 | 0.769146 | 0.728824 | 9 | GCAGGAAACCACAG | AAACCACAGAC | - |
| REL#MA0101.1 | -3 | 0.00322572 | 1.53544 | 0.728824 | 10 | GCAGGAAACCACAG | GGAAATCCCC | - |
| ETS1#MA0098.1 | -2 | 0.00377703 | 1.79787 | 0.728824 | 6 | GCAGGAAACCACAG | CGGAAA | - |
| Eip74EF#MA0026.1 | -1 | 0.00390936 | 1.86086 | 0.728824 | 7 | GCAGGAAACCACAG | CCGGAAG | + |
| SPIB#MA0081.1 | 0 | 0.00466848 | 2.2222 | 0.728824 | 7 | GCAGGAAACCACAG | AGAGGAA | + |
| GCR2#MA0305.1 | -2 | 0.00621779 | 2.95967 | 0.728824 | 7 | GCAGGAAACCACAG | AGGAAGC | - |
| En1#MA0027.1 | -3 | 0.00655102 | 3.11828 | 0.728824 | 11 | GCAGGAAACCACAG | GAACACTACTT | - |
| che-1#MA0260.1 | -4 | 0.00730751 | 3.47838 | 0.728824 | 6 | GCAGGAAACCACAG | GAAACC | + |
| ELF5#MA0136.1 | -1 | 0.00766575 | 3.6489 | 0.728824 | 9 | GCAGGAAACCACAG | AAGGAAGTA | - |
| Su(H)#MA0085.1 | 2 | 0.00973677 | 4.6347 | 0.833351 | 14 | GCAGGAAACCACAG | CTGTGGGAAACGAGAT | + |
| Tal1::Gata1#MA0140.1 | -1 | 0.0105182 | 5.00666 | 0.833351 | 13 | GCAGGAAACCACAG | CTGGTGGGGACAGATAAG | + |
| run::Bgb#MA0242.1 | -5 | 0.0129265 | 6.15302 | 0.883618 | 9 | GCAGGAAACCACAG | TAACCGCAA | + |
| Gamyb#MA0034.1 | -3 | 0.0130905 | 6.23108 | 0.883618 | 10 | GCAGGAAACCACAG | GACAACCGCC | + |
| dl_1#MA0022.1 | -1 | 0.0161719 | 7.69782 | 0.883618 | 12 | GCAGGAAACCACAG | CGGAAAAACCCC | - |
| Gfi#MA0038.1 | -4 | 0.0169461 | 8.06634 | 0.883618 | 10 | GCAGGAAACCACAG | CAAATCACTG | + |
| GCR1#MA0304.1 | -2 | 0.0180009 | 8.56844 | 0.883618 | 8 | GCAGGAAACCACAG | TGGAAGCC | + |
| SPI1#MA0080.2 | -2 | 0.018211 | 8.66842 | 0.883618 | 7 | GCAGGAAACCACAG | AGGAAGT | + |
| RELA#MA0107.1 | -3 | 0.0184392 | 8.77704 | 0.883618 | 10 | GCAGGAAACCACAG | GGAAATTCCC | - |
| RUNX1#MA0002.2 | -5 | 0.0185877 | 8.84776 | 0.883618 | 9 | GCAGGAAACCACAG | TAACCACAATA | - |