MacroAPE 1083:HSF1 f2
From FANTOM5_SSTAR
Full Name: HSF1_f2
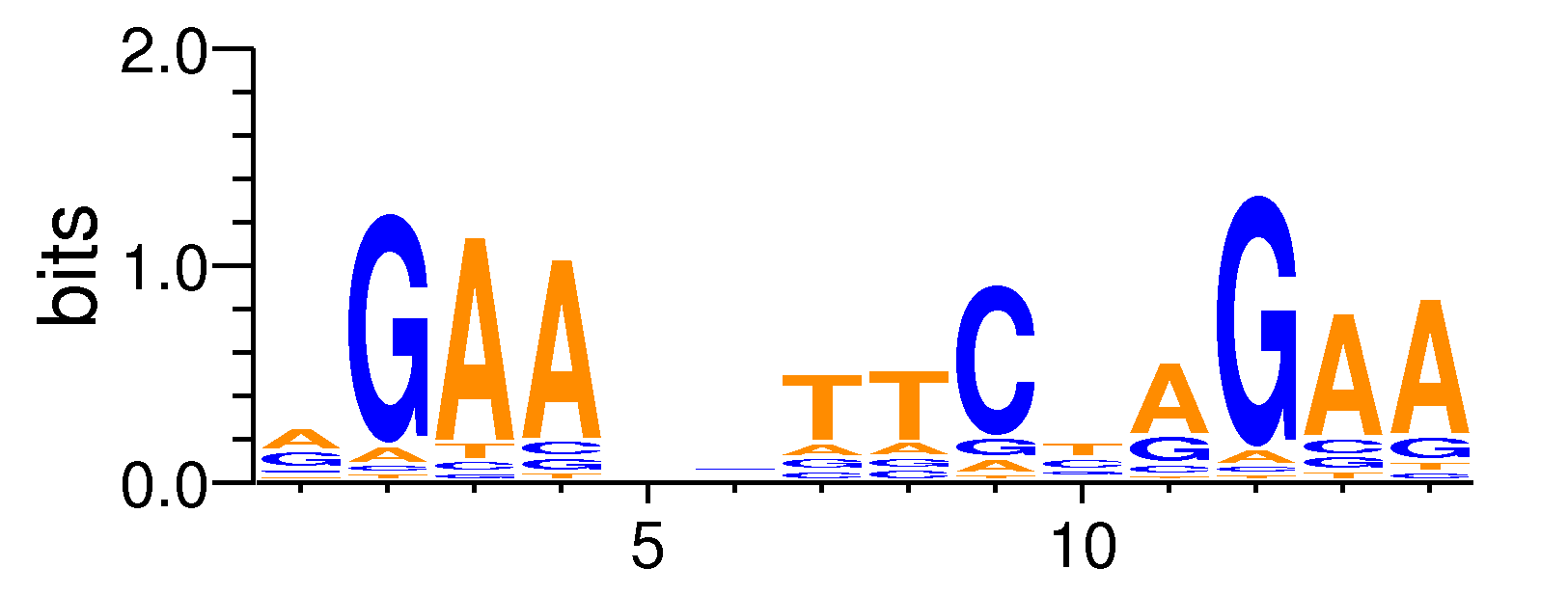
| ExpandMotif matrix |
|---|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| ABF2#MA0266.1 | -6 | 0.00215318 | 1.02491 | 1 | 7 | AGAAAGTTCTAGAA | CTCTAGA | + |
| MGA1#MA0336.1 | -2 | 0.00613697 | 2.9212 | 1 | 12 | AGAAAGTTCTAGAA | AGGACTATAGAACACTCTAAA | + |
| YER184C#MA0424.1 | -6 | 0.00938883 | 4.46908 | 1 | 8 | AGAAAGTTCTAGAA | CTCCGGAA | + |
| Cebpa#MA0102.1 | -2 | 0.0101259 | 4.81994 | 1 | 12 | AGAAAGTTCTAGAA | GAGATTGCGAAA | - |
| sd#MA0243.1 | -1 | 0.0105545 | 5.02394 | 1 | 12 | AGAAAGTTCTAGAA | GACATTCCTCGA | + |
| ARO80#MA0273.1 | 1 | 0.0188256 | 8.96099 | 1 | 14 | AGAAAGTTCTAGAA | GATAATTCTCGGCAAGTATGT | + |
| Stat3#MA0144.1 | -6 | 0.0202299 | 9.62943 | 1 | 8 | AGAAAGTTCTAGAA | TTCCAGGAAG | + |