MacroAPE 1083:Motif443
From FANTOM5_SSTAR
Full Name: motif443
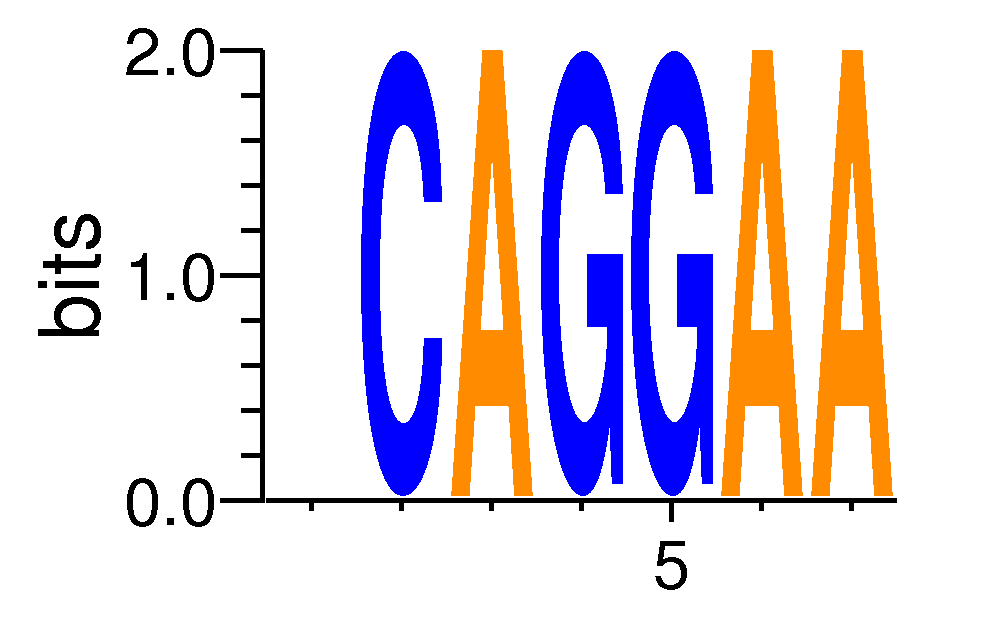
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| FEV#MA0156.1 | -1 | 0.000138771 | 0.0660552 | 0.0841472 | 6 | CCAGGAA | CAGGAAAT | + |
| Stat3#MA0144.1 | 2 | 0.000177763 | 0.0846153 | 0.0841472 | 7 | CCAGGAA | TTCCAGGAAG | + |
| Eip74EF#MA0026.1 | -1 | 0.001192 | 0.567394 | 0.303739 | 6 | CCAGGAA | CCGGAAG | + |
| TEAD1#MA0090.1 | 1 | 0.00128331 | 0.610856 | 0.303739 | 7 | CCAGGAA | CGGAGGAATGTG | - |
| SPIB#MA0081.1 | 0 | 0.00212971 | 1.01374 | 0.381148 | 7 | CCAGGAA | AGAGGAA | + |
| STAT1#MA0137.2 | 5 | 0.00241556 | 1.14981 | 0.381148 | 7 | CCAGGAA | GGTTTCCGGGAAATG | - |
| SPI1#MA0080.2 | -2 | 0.00379591 | 1.80685 | 0.42047 | 5 | CCAGGAA | AGGAAGT | + |
| ELF5#MA0136.1 | -1 | 0.00399715 | 1.90264 | 0.42047 | 6 | CCAGGAA | AAGGAAGTA | - |
| ELK1#MA0028.1 | 2 | 0.00479354 | 2.28172 | 0.453821 | 7 | CCAGGAA | GAGCCGGAAG | + |
| GABPA#MA0062.2 | 0 | 0.00606805 | 2.88839 | 0.478735 | 7 | CCAGGAA | ACCGGAAGAG | + |
| ASG1#MA0275.1 | -1 | 0.00932057 | 4.43659 | 0.635961 | 6 | CCAGGAA | CCGGAA | + |
| YBR239C#MA0420.1 | 0 | 0.00978868 | 4.65941 | 0.635961 | 7 | CCAGGAA | ACCGGAAC | + |
| LYS14#MA0325.1 | -1 | 0.0111718 | 5.31779 | 0.635961 | 6 | CCAGGAA | CCGGAATT | + |
| GABPA#MA0062.2 | -1 | 0.0121242 | 5.77111 | 0.635961 | 6 | CCAGGAA | CCGGAAGTGGC | + |
| MOT3#MA0340.1 | -1 | 0.0122295 | 5.82125 | 0.635961 | 6 | CCAGGAA | TAGGCA | + |
| ELK4#MA0076.1 | 0 | 0.0123375 | 5.87264 | 0.635961 | 7 | CCAGGAA | ACCGGAAGT | + |
| CAT8#MA0280.1 | -1 | 0.0137017 | 6.52202 | 0.635961 | 6 | CCAGGAA | CCGGAA | + |
| ETS1#MA0098.1 | -2 | 0.0142292 | 6.7731 | 0.635961 | 5 | CCAGGAA | CGGAAA | - |
| GCR2#MA0305.1 | -2 | 0.0147556 | 7.02367 | 0.635961 | 5 | CCAGGAA | AGGAAGC | - |
| SPI1#MA0080.2 | -2 | 0.0147783 | 7.03449 | 0.635961 | 5 | CCAGGAA | CGGAAG | + |
| IXR1#MA0323.1 | 2 | 0.0168269 | 8.00959 | 0.691903 | 7 | CCAGGAA | AAGCCGGAAGCGGGG | + |
| PUT3#MA0358.1 | 1 | 0.01754 | 8.34902 | 0.691903 | 7 | CCAGGAA | CCCGGGAA | + |