MacroAPE 1083:ZBT7A f1
From FANTOM5_SSTAR
Full Name: ZBT7A_f1
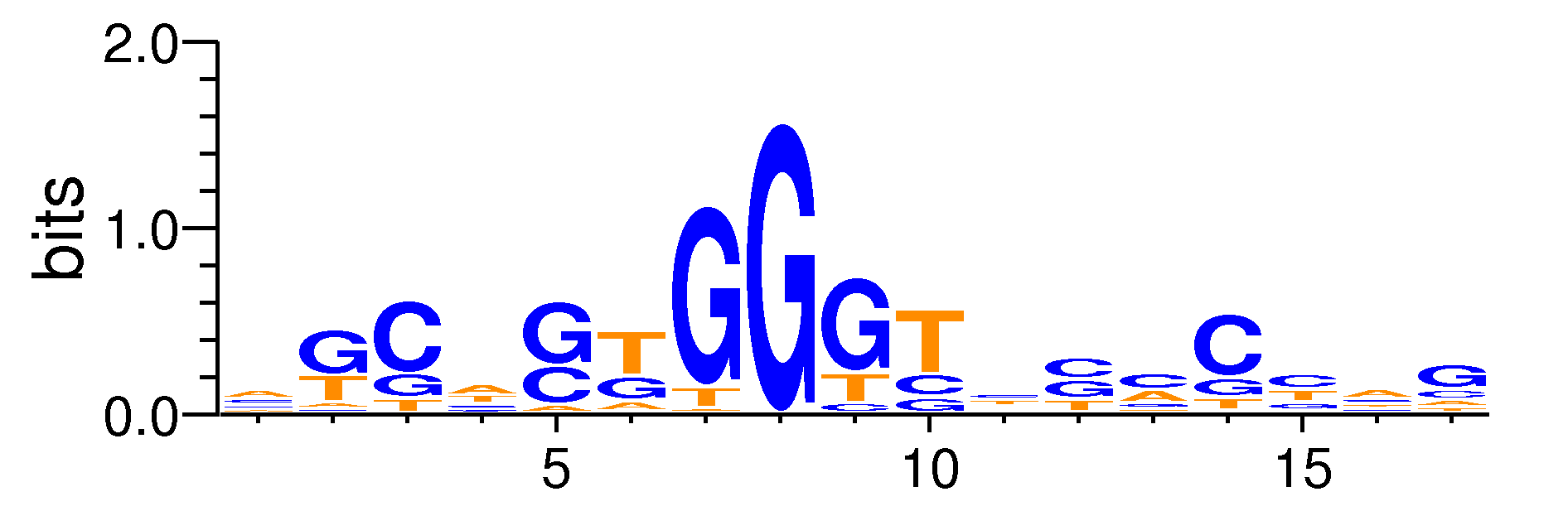
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| RUNX1#MA0002.2 | 0 | 0.000414622 | 0.19736 | 0.243924 | 11 | AGCAGTGGGTCCCCCAG | GTCTGTGGTTT | + |
| Pax4#MA0068.1 | 1 | 0.000518306 | 0.246714 | 0.243924 | 17 | AGCAGTGGGTCCCCCAG | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| Tal1::Gata1#MA0140.1 | -4 | 0.00132681 | 0.63156 | 0.259751 | 13 | AGCAGTGGGTCCCCCAG | CTGGTGGGGACAGATAAG | + |
| NRG1#MA0347.1 | 2 | 0.00135223 | 0.643661 | 0.259751 | 17 | AGCAGTGGGTCCCCCAG | CTAGATCAGGGTCCATCGCA | + |
| opa#MA0456.1 | 1 | 0.00137984 | 0.656804 | 0.259751 | 11 | AGCAGTGGGTCCCCCAG | CAGCGGGGGGTC | - |
| Macho-1#MA0118.1 | -2 | 0.00271024 | 1.29007 | 0.425163 | 9 | AGCAGTGGGTCCCCCAG | TGGGGGGTC | + |
| NFKB1#MA0105.1 | -4 | 0.00358783 | 1.70781 | 0.482428 | 11 | AGCAGTGGGTCCCCCAG | GGGGATTCCCC | + |
| YGR067C#MA0425.1 | 3 | 0.0041964 | 1.99749 | 0.493726 | 11 | AGCAGTGGGTCCCCCAG | TGAAAAAGTGGGGT | - |
| PPARG#MA0066.1 | -3 | 0.0055471 | 2.64042 | 0.565082 | 14 | AGCAGTGGGTCCCCCAG | AGTAGGTCACCGTGACCTAC | - |
| YPR022C#MA0436.1 | -3 | 0.00600361 | 2.85772 | 0.565082 | 7 | AGCAGTGGGTCCCCCAG | CGTGGGG | - |
| ESR1#MA0112.2 | 0 | 0.00680533 | 3.23934 | 0.582312 | 17 | AGCAGTGGGTCCCCCAG | AGGTCAGGGTGACCTGGGCC | - |
| AFT1#MA0269.1 | 0 | 0.0099228 | 4.72325 | 0.655732 | 17 | AGCAGTGGGTCCCCCAG | CCAATCGGGTGCAATATGTAA | - |
| MET28#MA0332.1 | -2 | 0.0111386 | 5.30198 | 0.655732 | 6 | AGCAGTGGGTCCCCCAG | CTGTGG | + |
| ADR1#MA0268.1 | -4 | 0.0113116 | 5.38432 | 0.655732 | 7 | AGCAGTGGGTCCCCCAG | GTGGGGT | - |
| YML081W#MA0431.1 | -2 | 0.0113896 | 5.42143 | 0.655732 | 9 | AGCAGTGGGTCCCCCAG | GTGCGGGGT | - |
| MIG1#MA0337.1 | -4 | 0.0120962 | 5.75778 | 0.655732 | 7 | AGCAGTGGGTCCCCCAG | GCGGGGG | - |
| ZNF354C#MA0130.1 | -4 | 0.0127788 | 6.08269 | 0.655732 | 6 | AGCAGTGGGTCCCCCAG | GTGGAT | - |
| CRZ1#MA0285.1 | -4 | 0.013618 | 6.48219 | 0.655732 | 9 | AGCAGTGGGTCCCCCAG | GTGGCTTAG | - |
| DOT6#MA0351.1 | 4 | 0.0139334 | 6.6323 | 0.655732 | 17 | AGCAGTGGGTCCCCCAG | AGGATGCGATGAGGTGCAGAA | - |
| Tcfcp2l1#MA0145.1 | -4 | 0.0153831 | 7.32235 | 0.689482 | 13 | AGCAGTGGGTCCCCCAG | CTGGTTTGAACTGG | - |
| Gfi#MA0038.1 | -2 | 0.0169317 | 8.05949 | 0.701969 | 10 | AGCAGTGGGTCCCCCAG | CAGTGATTTG | - |
| STP2#MA0395.1 | 1 | 0.0171949 | 8.18479 | 0.701969 | 17 | AGCAGTGGGTCCCCCAG | TCGTCGTGCGGCGCCGATCA | - |
| dl_1#MA0022.1 | -4 | 0.0201386 | 9.58595 | 0.706403 | 12 | AGCAGTGGGTCCCCCAG | GGGGTTTTTCCC | + |