Novel motif:91
From FANTOM5_SSTAR
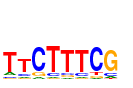
| name: | motif91_TTCTTTCG |
|---|
Association to promoter expression in human samples<b>Summary:</b>Significance of the correlation with CAGE expression, human <br><b>Analyst:</b> Michiel de Hoon<br><br>link to source dataset <br>data
Association to promoter expression in mouse samples
<b>Summary:</b>Significance of the correlation with CAGE expression, mouse <br><b>Analyst:</b> Michiel de Hoon<br><br>link to source dataset <br>data
GREAT analysis results for human
<b>Summary:</b>Genomic Enrichment of Annotations Tool (GREAT, Nat Biotechnol. 2010 May;28(5):495-501) is used to identify, both in human and in mouse, the gene ontology terms of biological processes enriched given the predicted TFBSs, and evaluated the overlap in the top-500 gene ontology terms between human and mouse. For each novel motif, the P value for the overlap was then evaluated by calculating its relative rank with respect to this background distribution.<br>Analyst: Michiel de Hoon<br><br>link to source dataset <br>data
GREAT analysis results for mouseGenomic Enrichment of Annotations Tool (GREAT, Nat Biotechnol. 2010 May;28(5):495-501) is used to identify, both in human and in mouse, the gene ontology terms of biological processes enriched given the predicted TFBSs, and evaluated the overlap in the top-500 gene ontology terms between human and mouse. For each novel motif, the P value for the overlap was then evaluated by calculating its relative rank with respect to this background distribution.<br>Analyst: Michiel de Hoon <br><br>link to source dataset <br>data