MacroAPE 1083:SRF do
From FANTOM5_SSTAR
Full Name: SRF_do
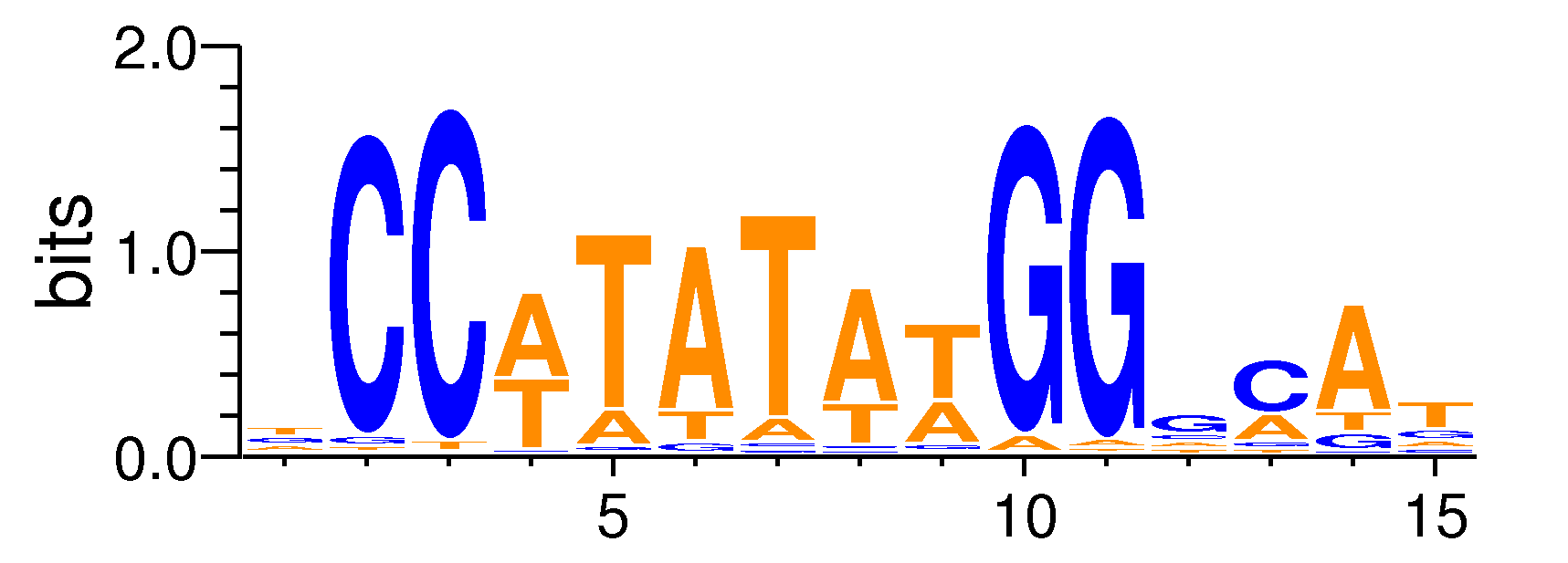
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SRF#MA0083.1 | -1 | 2.63783e-12 | 1.25561e-09 | 2.49202e-09 | 12 | TCCATATATGGGCAT | CCATATATGGGC | - |
| AG#MA0005.1 | 0 | 1.25236e-05 | 0.00596121 | 0.00260267 | 11 | TCCATATATGGGCAT | ACCAAATTTGG | - |
| squamosa#MA0082.1 | -1 | 1.27607e-05 | 0.00607411 | 0.00260267 | 14 | TCCATATATGGGCAT | CCAAAAATGGAAAT | + |
| AGL3#MA0001.1 | -1 | 1.37747e-05 | 0.00655678 | 0.00260267 | 10 | TCCATATATGGGCAT | CTATTTATGG | - |
| MCM1#MA0331.1 | 1 | 0.00183275 | 0.872389 | 0.192383 | 11 | TCCATATATGGGCAT | TTCCTAATTGGG | - |
| TBP#MA0108.2 | -1 | 0.00249126 | 1.18584 | 0.213959 | 14 | TCCATATATGGGCAT | GTATAAAAGGCGGGG | + |
| TBP#MA0108.2 | -1 | 0.00249126 | 1.18584 | 0.213959 | 14 | TCCATATATGGGCAT | GTATAAAAGGCGGGG | + |
| Hltf#MA0109.1 | -3 | 0.00555203 | 2.64277 | 0.403472 | 10 | TCCATATATGGGCAT | ATATAAGGTT | - |
| NHP6B#MA0346.1 | 4 | 0.00621895 | 2.96022 | 0.419656 | 15 | TCCATATATGGGCAT | TCATATTATATATAAATAAA | - |
| ASH1#MA0276.1 | -1 | 0.00758793 | 3.61186 | 0.4779 | 10 | TCCATATATGGGCAT | CCCGATCCGG | - |
| NHP6A#MA0345.1 | 4 | 0.00832536 | 3.96287 | 0.488001 | 15 | TCCATATATGGGCAT | ATGACCTATATATAAAAATGA | + |
| YER130C#MA0423.1 | -4 | 0.0101818 | 4.84652 | 0.506261 | 9 | TCCATATATGGGCAT | AATAGGGGT | - |
| SIG1#MA0379.1 | -4 | 0.01237 | 5.88812 | 0.556488 | 5 | TCCATATATGGGCAT | TATAT | - |
| Gata1#MA0035.2 | 0 | 0.0163276 | 7.77194 | 0.67235 | 11 | TCCATATATGGGCAT | TTCTTATCTGT | - |
| twi#MA0249.1 | -1 | 0.0163689 | 7.79158 | 0.67235 | 12 | TCCATATATGGGCAT | CAACATATGCGA | - |