MacroAPE 1083:GGGTTGCGTCA,motif101
From FANTOM5_SSTAR
Full Name: GGGTTGCGTCA,motif101,CNhs13473,T:2494.0(41.24%),B:25191.5(31.65%),P:1e-54
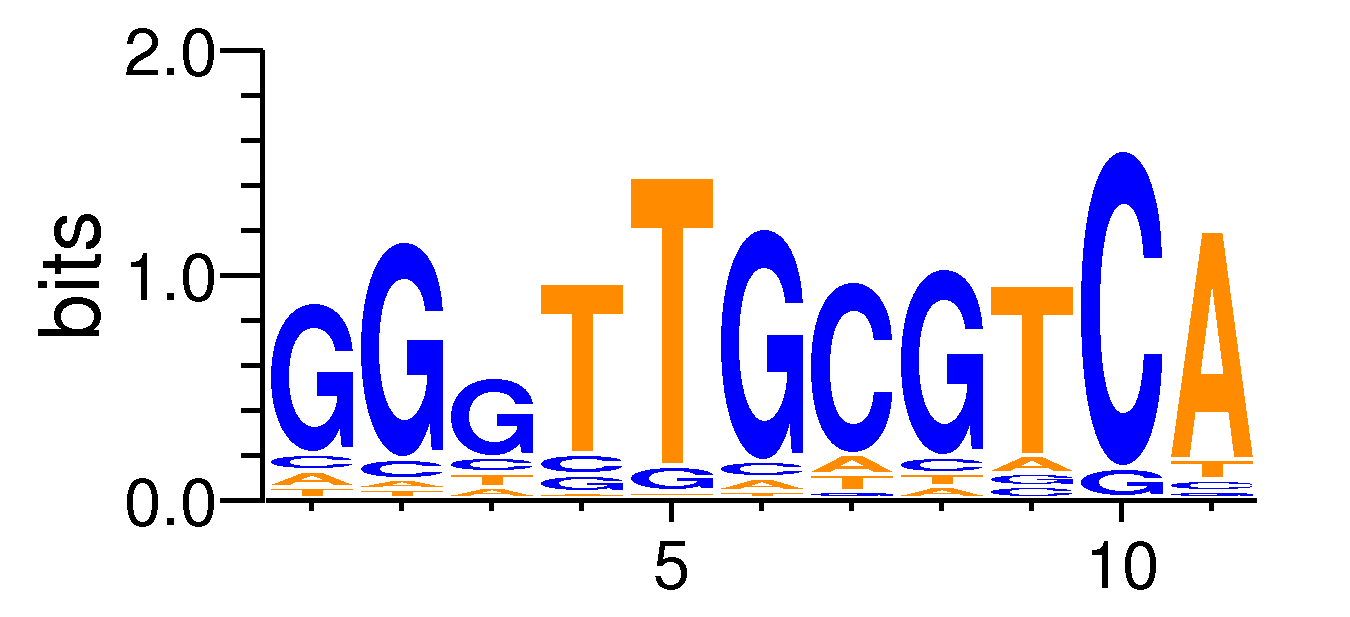
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| FHL1#MA0295.1 | -2 | 0.000159952 | 0.0761373 | 0.151111 | 8 | GGGTTGCGTCA | TTTGCGTC | - |
| Cebpa#MA0102.1 | 1 | 0.000997785 | 0.474945 | 0.352332 | 11 | GGGTTGCGTCA | GAGATTGCGAAA | - |
| AP1#MA0099.2 | -4 | 0.00161513 | 0.768803 | 0.352332 | 7 | GGGTTGCGTCA | TGAGTCA | - |
| ARG80#MA0271.1 | -5 | 0.00204647 | 0.974119 | 0.352332 | 6 | GGGTTGCGTCA | GCGTCT | - |
| CREB1#MA0018.2 | -3 | 0.00214744 | 1.02218 | 0.352332 | 8 | GGGTTGCGTCA | TGACGTCA | - |
| CEBPA#MA0102.2 | -2 | 0.00245288 | 1.16757 | 0.352332 | 9 | GGGTTGCGTCA | ATTGCGAAA | - |
| CIN5#MA0284.1 | -1 | 0.00284204 | 1.35281 | 0.352332 | 10 | GGGTTGCGTCA | GATTACGTAA | - |
| BRCA1#MA0133.1 | 0 | 0.0033031 | 1.57227 | 0.352332 | 7 | GGGTTGCGTCA | GTGTTGT | - |
| TGA1A#MA0129.1 | -4 | 0.00403898 | 1.92256 | 0.352332 | 7 | GGGTTGCGTCA | TACGTCA | + |
| RPN4#MA0373.1 | -1 | 0.00424246 | 2.01941 | 0.352332 | 7 | GGGTTGCGTCA | GGTGGCG | + |
| CUP9#MA0288.1 | -2 | 0.00428629 | 2.04027 | 0.352332 | 9 | GGGTTGCGTCA | AATGTGTCA | - |
| HLF#MA0043.1 | -1 | 0.00447537 | 2.13028 | 0.352332 | 10 | GGGTTGCGTCA | GGTTACGCAATT | + |
| CST6#MA0286.1 | -3 | 0.00540189 | 2.5713 | 0.364521 | 8 | GGGTTGCGTCA | TTACGTCAT | - |
| Ddit3::Cebpa#MA0019.1 | 1 | 0.0058484 | 2.78384 | 0.368341 | 11 | GGGTTGCGTCA | GGGATTGCATCT | - |
| GCN4#MA0303.1 | 3 | 0.0109614 | 5.21761 | 0.647216 | 11 | GGGTTGCGTCA | CAAGGGATGAGTCATACTTCA | + |
| AFT2#MA0270.1 | 0 | 0.0119997 | 5.71185 | 0.666847 | 8 | GGGTTGCGTCA | GGGGTGTG | - |
| NFIL3#MA0025.1 | 0 | 0.0148742 | 7.0801 | 0.699865 | 11 | GGGTTGCGTCA | ATGTTACATAA | - |
| SNT2#MA0384.1 | 0 | 0.0153355 | 7.2997 | 0.699865 | 11 | GGGTTGCGTCA | TGGTAGCGCCG | + |
| gt#MA0447.1 | -2 | 0.0162544 | 7.73712 | 0.699865 | 9 | GGGTTGCGTCA | ATTACGTAAT | + |
| NFE2L2#MA0150.1 | -1 | 0.0162979 | 7.75781 | 0.699865 | 10 | GGGTTGCGTCA | TGCTGAGTCAT | - |
| CREB1#MA0018.2 | 2 | 0.0175208 | 8.3399 | 0.719666 | 10 | GGGTTGCGTCA | CCTGGTGACGTC | + |
| Spz1#MA0111.1 | 1 | 0.0201235 | 9.5788 | 0.792132 | 10 | GGGTTGCGTCA | AGGGTAACAGC | + |