MacroAPE 1083:NFIX
From FANTOM5_SSTAR
Full Name: NFIX
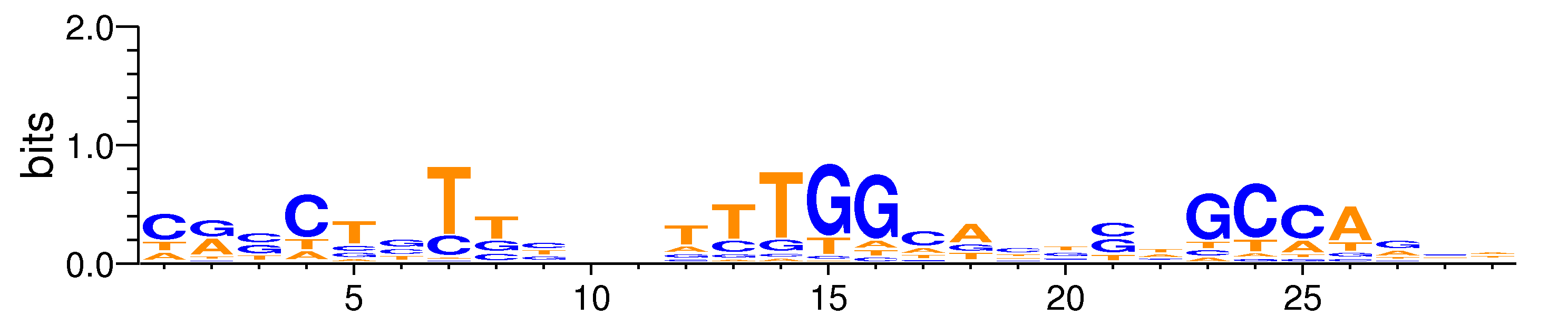
| Motif matrix | |
|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| TLX1::NFIC#MA0119.1 | -12 | 1.38522e-09 | 6.59363e-07 | 1.29662e-06 | 14 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTGGCATGGTGCCA | - |
| NFIC#MA0161.1 | -12 | 3.28456e-06 | 0.00156345 | 0.00103899 | 6 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTGGCA | + |
| STP2#MA0395.1 | -9 | 0.000501178 | 0.238561 | 0.117281 | 20 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TGATCGGCGCCGCACGACGA | + |
| Hand1::Tcfe2a#MA0092.1 | -9 | 0.000716586 | 0.341095 | 0.118572 | 10 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GGTCTGGCAT | + |
| HAP4#MA0315.1 | -2 | 0.00109177 | 0.519681 | 0.1375 | 15 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TCGCCTCTGATTGGT | - |
| Pax4#MA0068.1 | -3 | 0.00117516 | 0.559376 | 0.1375 | 26 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| BAS1#MA0278.1 | -7 | 0.00132965 | 0.632912 | 0.138289 | 21 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTGACTTGACTCTGGCTGTGC | - |
| HAP2#MA0313.1 | -12 | 0.00153362 | 0.730005 | 0.143553 | 5 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTGGT | + |
| NFYA#MA0060.1 | -5 | 0.00221747 | 1.05551 | 0.175298 | 16 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GCGCTGATTGGCTGAG | - |
| AFT1#MA0269.1 | -6 | 0.00232851 | 1.10837 | 0.175298 | 21 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTACATATTGCACCCGATTGG | + |
| HMG-I/Y#MA0045.1 | -2 | 0.00246038 | 1.17114 | 0.175298 | 16 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GTTTTTCCATTTGTTG | - |
| HAP5#MA0316.1 | -2 | 0.00262187 | 1.24801 | 0.175298 | 15 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TGGGATCTGATTGGT | + |
| id1#MA0120.1 | -4 | 0.00283691 | 1.35037 | 0.177031 | 12 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTTTCCTTTTCG | + |
| exd#MA0222.1 | -10 | 0.00416171 | 1.98097 | 0.229739 | 8 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTTTGACA | + |
| STB3#MA0390.1 | -3 | 0.00417244 | 1.98608 | 0.229739 | 21 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GTCCAAAATTTTTCACTCTGG | + |
| NDT80#MA0343.1 | -2 | 0.00475725 | 2.26445 | 0.247388 | 21 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTGCGTTTTTGTGGCCGGAAA | - |
| E2F1#MA0024.1 | -11 | 0.00548556 | 2.61113 | 0.270248 | 8 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTTGGCGC | + |
| ARO80#MA0273.1 | -4 | 0.00590302 | 2.80984 | 0.276273 | 21 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GATAATTCTCGGCAAGTATGT | + |
| SUT2#MA0400.1 | -6 | 0.00912356 | 4.34282 | 0.379734 | 20 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | AAATTTTCGGAGTTTATCAT | - |
| TOS8#MA0408.1 | -11 | 0.00933066 | 4.44139 | 0.379734 | 8 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTTGACAG | - |
| Foxa2#MA0047.2 | -4 | 0.0107563 | 5.12 | 0.419514 | 12 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TGTTTACTTAGG | + |
| AG#MA0005.1 | -5 | 0.0113344 | 5.39519 | 0.423121 | 11 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | ACCAAATTTGG | - |
| YAP6#MA0418.1 | -6 | 0.0117528 | 5.59435 | 0.423121 | 20 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GTCGCTTACGTAATCAGACC | - |
| SFL1#MA0377.1 | 3 | 0.0124715 | 5.93645 | 0.432365 | 18 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTTTATTTCTTCTATTCTCTA | - |
| RIM101#MA0368.1 | -11 | 0.0135621 | 6.45556 | 0.447747 | 7 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | CTTGGCG | - |
| RTG3#MA0376.1 | -9 | 0.0138719 | 6.60302 | 0.447747 | 20 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | ATATTAGCACGTGCTTAGTC | + |
| Cebpa#MA0102.1 | -11 | 0.0148173 | 7.05304 | 0.456039 | 12 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TTTCGCAATCTT | + |
| Tcfcp2l1#MA0145.1 | -2 | 0.0151032 | 7.18912 | 0.456039 | 14 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | CTGGTTTGAACTGG | - |
| HNF4A#MA0114.1 | -6 | 0.0157343 | 7.48951 | 0.456967 | 13 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TGACCTTTGGCCT | - |
| MET31#MA0333.1 | -9 | 0.0164639 | 7.83681 | 0.456967 | 9 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | GGTGTGGCG | + |
| NHP6A#MA0345.1 | -3 | 0.0171413 | 8.15928 | 0.456967 | 21 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | ATGACCTATATATAAAAATGA | + |
| Tal1::Gata1#MA0140.1 | -9 | 0.0175749 | 8.36564 | 0.456967 | 18 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | CTTATCTGTCCCCACCAG | - |
| FOXA1#MA0148.1 | -4 | 0.0209923 | 9.99232 | 0.531071 | 11 | CGCCTGTTCTTTTTGGCACTCTGCCAGCA | TGTTTACTTTG | + |