Genetic information in the genome is used as blueprint for protein construction, by being transcribed to mRNA and then translated into the amino acid sequences that constitute the protein. Since the amino acid sequences are directly coded in the base sequence of the mRNA, analysis of the mRNA sequence is very important for analysis of protein function. In the “Mouse encyclopedia project”, we gathered a comprehensive set of cDNA clones – which are complete, full-length copies of the mRNAs – from various tissues and cell types. Here, we will give you a general description of the full-length cDNA libraries developed by our research group. These technologies are now being used to analyze cDNAs of various biological species.
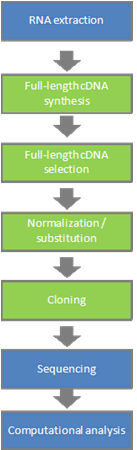
Technical overview of the full-length cDNA project
Key technologies
- Trehalose method[1]
Until now, the biggest problem of reverse transcriptional response has been the inefficiency in the synthesis of the first cDNA strand. We discovered that this problem can be solved by adding trehalose. It was previously known that trehalose can be induced by adding yeast, which is treated by heat-shock using disaccharides. This fact was a hint that we used to discover that trehalose gives heat resistance to enzyme including the reverse transcriptional enzyme. From this discovery, it became possible to induce reactions using the reverse transcriptional enzyme with 60°C instead of 42°C as was previously the norm. Under 60°C, the template RNAs' secondary structure gets weaker, and the area with stable secondary structure, which often exists in non-coding 5' end mRNAs, becomes possible to be transcribed in reverse in a very efficient manner. - Cap-trapper method[2] [3]
We developed the biotynylated-cap-trapper technique to remove cDNAs whose elongation was ended incompletely from the libraries.
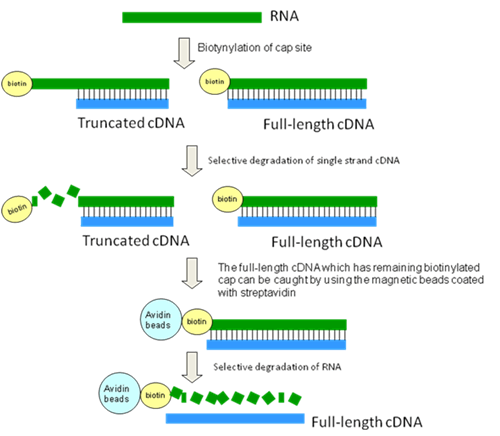
With this method, we biotinylate the cap-structure that is specific to mRNA of eukaryotic organisms. We then selectively extract the biotinylated cap structure of non full-length cDNA. The remaining full-length cDNA containing a biotinylated cap can be caught by using magnetic beads coated with streptavidin. The selected full-length cDNA can then be eluted by alkaline treatment. Synthesizing the second strand from the 1st strand cDNA (which became single stranded when separated from the beads) makes it possible to obtain the full-length cDNA selectively. - Novel cloning vector[4]
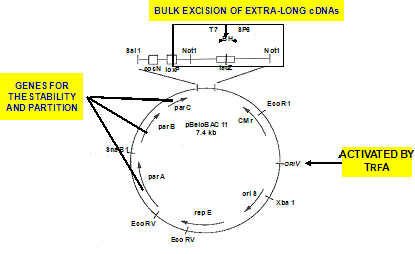
In the past, there was a problem called "cloning-bias" in the production of full-length cDNA libraries. Clones with shorter mRNAs are more likely to be cloned, and short clones are enriched. In addition to the fact that cDNA can be produced more easily compared to long cDNA in the presence of trehalose, ordinary vectors also have a higher tendency to clone short cDNA compared to long cDNA. Our research group started to produce a new vector capable of covering cDNA spanning 6kb~20kb. This new vector, named "λFlcIII-L", gave an average strand length of 6.9kb, the longest among existing cDNA libraries. We produced λFlcIV from the improvement of the new vector "λFlcIII-L" to decrease background, and also to apply to the functional analysis after the determination of sequences. With λFlcIV, we managed to produce cDNA of with almost zero background.
- Normalisation and substitution[5]
50~60% of all RNA is house keeping mRNAs. When trying to identify cDNA with very low expression levels (down to one sequence), the identification efficiency gets considerably low. This became a serious obstacle for our project, where the aim was to identify all expressed RNA comprehensively. In order to improve the efficiency of random sequence analysis, it is necessary to reduce the proportion of cDNA that is highly expressed in the libraries. In general terms, all identified cDNA can be deleted from libraries. To do this, we established the "cDNA normalisation" method to homogenize the frequency of cDNA and the "subtraction" method to delete cloned cDNA from libraries. In the dynamics of the re-engagement of single strand nucleotides to the complementary strand, nucleotides with higher concentration generally re-engage quicker than the ones with lower concentration. Both normalisation and s subtraction is utilize this phenomenon. First, mRNA that is left from the preparation of cDNA get biotynilated (Driver), then the same amount of driver as the first cDNA strand (it is single strand DNA, and the sequence is complementary sequence to mRNA, hence it is called "tester") is mixed in. Under the appropriate control (especially to induce re-engagement of high to middle frequencies), since biotynilated mRNA is attached, cDNA that is re-engaged with mRNA can be removed selectively by using the magnetic beads.
Applications
A series of cDNA technologies has been improved to become applicable to very small tissues, and are now applied to various biological species. We have developed CAGE as the applied technology of the Cap-trapper technique, where the 5' end of mRNA is cut off as a tag. By utilizing these techniques we have achieved significant success, such as the discovery of the "RNA continent".
References
- ^ P. Carninci, Y. Nishiyama, A. Westover, M. Itoh, S. Nagaoka, N. Sasaki, Y. Okazaki, M. Muramatsu, and Y. Hayashizaki, Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA, Proc Natl Acad Sci U S A 95, 520-524 (1998), doi: 10.1073/pnas.95.2.520
- ^ P. Carninci, C. Kvam, A. Kitamura, T. Ohsumi, Y. Okazaki, M. Itoh, M. Kamiya, K. Shibata, N. Sasaki, M. Izawa, M. Muramatsu, Y. Hayashizaki, and C. Schneider, High-efficiency full-length cDNA cloning by biotinylated CAP trapper, Genomics 37, 327-336 (1996), doi; 10.1006/geno.1996.0567
- ^ P. Carninci, A. Westover, Y. Nishiyama, T. Ohsumi, M. Itoh, S. Nagaoka, N. Sasaki, Y. Okazaki, M. Muramatsu, C. Schneider, and Y. Hayashizaki, High efficiency selection of full-length cDNA by improved biotinylated cap trapper, DNA Res 4, 61-66 (1997), doi: 10.1093/dnares/4.1.61
- ^ P. Carninci, Y. Shibata, N. Hayatsu, M. Itoh, T. Shiraki, T. Hirozane, A. Watahiki, K. Shibata, H. Konno, M. Muramatsu, and Y. Hayashizaki, Balanced-size and long-size cloning of full-length, cap-trapped cDNAs into vectors of the novel lambda-FLC family allows enhanced gene discovery rate and functional analysis, Genomics 77, 79-90 (2001), doi: 10.1006/geno.2001.6601
- ^ P. Carninci, Y. Shibata, N. Hayatsu, Y. Sugahara, K. Shibata, M. Itoh, H. Konno, Y. Okazaki, M. Muramatsu, and Y. Hayashizaki, Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes, Genome Res 10, 1617-1630 (2000), doi: 10.1006/geno.2001.6601
